5RYO
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z136583524 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-N-(pyridin-4-yl)furan-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5R4E
 
 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with RZ201 | | Descriptor: | (5R)-5-[(4-fluorophenyl)methyl]-5-(2-hydroxyethyl)-3-(2-methoxyethyl)imidazolidine-2,4-dione, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
5RYX
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2234920345 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZE
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434786 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5R9V
 
 | |
5RZX
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434814 | | Descriptor: | 1,2-ETHANEDIOL, 3-(diethylamino)phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RAB
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001726a | | Descriptor: | (2R,5S)-5-(4-chlorophenyl)oxolane-2-carbohydrazide, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAS
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with XS036302b | | Descriptor: | 2-(4-phenylpiperidin-1-yl)ethanoic acid, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5REB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2856434899 | | Descriptor: | 1-[(thiophen-3-yl)methyl]piperidin-4-ol, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5R53
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13854a | | Descriptor: | DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22, ~{N}-[(3~{R})-2-oxidanylideneazepan-3-yl]furan-2-carboxamide | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RGO
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102248 (Mpro-x0736) | | Descriptor: | 1-[4-(furan-2-carbonyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5R5H
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13706a | | Descriptor: | 3-chloro-N-(1-hydroxy-2-methylpropan-2-yl)benzamide, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RH1
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2010253653 (Mpro-x2643) | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5R5U
 
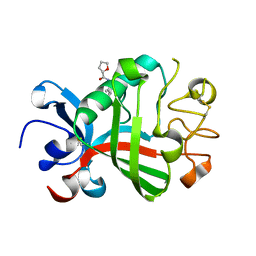 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z1545312521 | | Descriptor: | (2S)-N-(3-chloro-2-methylphenyl)oxolane-2-carboxamide, Tenascin C (Hexabrachion), isoform CRA_a | | Authors: | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RHG
 
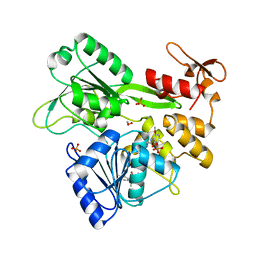 | |
5R7W
 
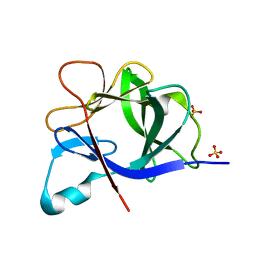 | |
5RHW
 
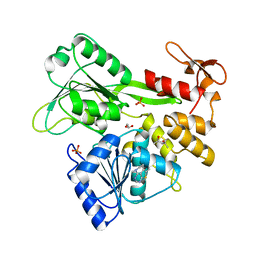 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z31222641 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, NS3 Helicase, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R86
 
 | |
5RJT
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z404993336 | | Descriptor: | 5-methyl-N-(1-methyl-1H-pyrazol-4-yl)-1,2-oxazole-3-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.169 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5R8H
 
 | |
5RK8
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z53116498 | | Descriptor: | 3-(2-methyl-1H-benzimidazol-1-yl)propanamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z373768898 | | Descriptor: | N-(1-ethyl-1H-pyrazol-4-yl)cyclopentanecarboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5R8Y
 
 | |
5RKZ
 
 | | PanDDA analysis group deposition of ground-state model of human NUDT22 screened against the DSPL fragment library by X-ray Crystallography | | Descriptor: | DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RLF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z235341991 | | Descriptor: | Helicase, N-(2-methoxy-5-methylphenyl)glycinamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
