1GNM
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASP (V82D) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
1GNN
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASN (V82N) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
1GNO
 
 | | HIV-1 PROTEASE (WILD TYPE) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
1GNP
 
 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID 3'-O-(N-METHYLANTHRANILOYL-2'-DEOXYGUANYLATE ESTER | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
1GNQ
 
 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
1GNR
 
 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
1GNS
 
 | | SUBTILISIN BPN' | | Descriptor: | ACETONE, SUBTILISIN BPN' | | Authors: | Almog, O, Gallagher, D.T, Ladner, J.E, Strausberg, S, Alexander, P. | | Deposit date: | 2001-10-06 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Thermostability. Analysis of Stabilizing Mutations in Subtilisin Bpn'.
J.Biol.Chem., 277, 2002
|
|
1GNT
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris. X-ray structure at 1.25A resolution using synchrotron radiation. | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 A resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 7, 2002
|
|
1GNU
 
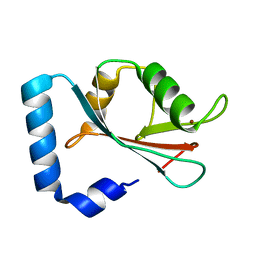 | | GABA(A) receptor associated protein GABARAP | | Descriptor: | GABARAP, NICKEL (II) ION | | Authors: | Knight, D, Harris, R, Moss, S, Driscoll, P.C, Keep, N.H. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Crystal Structure and Putative Ligand-Derived Peptide Binding Properties of Gamma-Aminobutyric Acid Receptor Type a Receptor-Associated Protein
J.Biol.Chem., 277, 2002
|
|
1GNV
 
 | |
1GNW
 
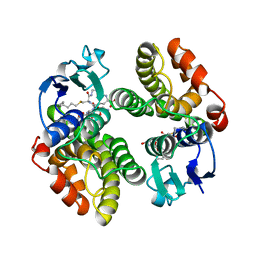 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Reinemer, P, Prade, L, Hof, P, Neuefeind, T, Huber, R, Palme, K, Bartunik, H.D, Bieseler, B. | | Deposit date: | 1996-09-15 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of glutathione S-transferase from Arabidopsis thaliana at 2.2 A resolution: structural characterization of herbicide-conjugating plant glutathione S-transferases and a novel active site architecture.
J.Mol.Biol., 255, 1996
|
|
1GNX
 
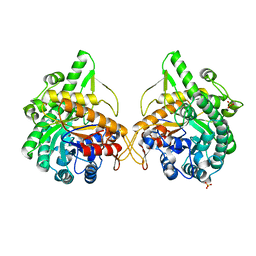 | | b-glucosidase from Streptomyces sp | | Descriptor: | BETA-GLUCOSIDASE, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Guasch, A, Perez-Pons, J.A, Vallmitjana, M, Querol, E, Coll, M. | | Deposit date: | 2001-10-10 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Beta-Glucosidase from Streptomyces
To be Published
|
|
1GNY
 
 | | xylan-binding module CBM15 | | Descriptor: | SODIUM ION, XYLANASE 10C, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Szabo, S, Jamal, S, Xie, H, Charnock, S.J, Bolam, D.N, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of a Family 15 Carbohydrate-Binding Module in Complex with Xylopentaose: Evidence that Xylan Binds in an Approximate Three-Fold Helical Conformation
J.Biol.Chem., 276, 2001
|
|
1GNZ
 
 | | LECTIN I-B4 FROM GRIFFONIA SIMPLICIFOLIA (GS I-B4)METAL FREE FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GSI-B4 ISOLECTIN, PHOSPHATE ION | | Authors: | Lescar, J, Loris, R, Mitchell, E, Gautier, C, Imberty, A. | | Deposit date: | 2001-10-11 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isolectins I-A and I-B of Griffonia (Bandeiraea) Simplicifolia. Crystal Structure of Metal-Free Gs I-B(4) and Molecular Basis for Metal Binding and Monosaccharide Specificity.
J.Biol.Chem., 277, 2002
|
|
1GO0
 
 | |
1GO1
 
 | |
1GO2
 
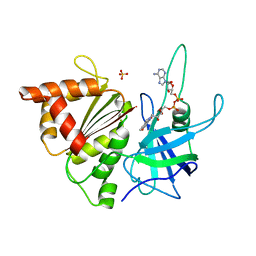 | | Structure of Ferredoxin-NADP+ Reductase with Lys 72 replaced by Glu (K72E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-10-15 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners
Proteins: Struct.,Funct., Genet., 59, 2005
|
|
1GO3
 
 | | Structure of an archeal homolog of the eukaryotic RNA polymerase II RPB4/RPB7 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE SUBUNIT E, DNA-DIRECTED RNA POLYMERASE SUBUNIT F | | Authors: | Todone, F, Brick, P, Werner, F, Weinzierl, R.O.J, Onesti, S. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an Archaeal Homolog of the Eukaryotic RNA Polymerase II Rpb4/Rpb7 Complex
Mol.Cell, 8, 2001
|
|
1GO4
 
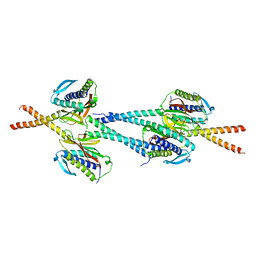 | | Crystal structure of Mad1-Mad2 reveals a conserved Mad2 binding motif in Mad1 and Cdc20. | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD1, Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Sironi, L, Mapelli, M, Jeang, K.T, Musacchio, A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint.
EMBO J., 21, 2002
|
|
1GO5
 
 | |
1GO6
 
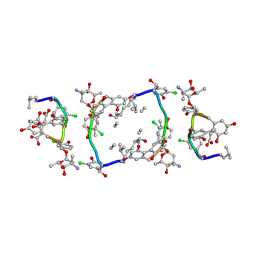 | | Balhimycin in complex with Lys-D-ala-D-ala | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Lehmann, C, Bunkoczi, G, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2001-10-19 | | Release date: | 2002-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
1GO7
 
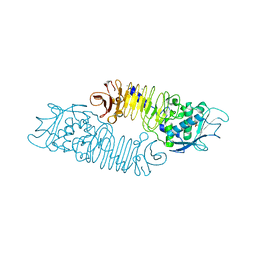 | | The metzincin's methionine: PrtC M226C-E189K double mutant | | Descriptor: | CALCIUM ION, PHOSPHATE ION, PROTEASE C, ... | | Authors: | Hege, T. | | Deposit date: | 2001-10-20 | | Release date: | 2002-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Complex between Pseudomonas Aeruginosa Alkaline Protease and its Cognate Inhibitor: Inhibition by a Zinc-NH2 Coordinative Bond
J.Biol.Chem., 276, 2001
|
|
1GO8
 
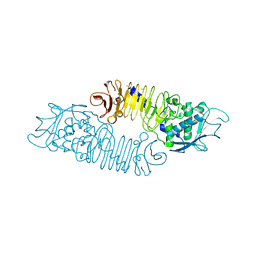 | | The metzincin's methionine: PrtC M226L mutant | | Descriptor: | CALCIUM ION, PHOSPHATE ION, PROTEASE C, ... | | Authors: | Baumann, U. | | Deposit date: | 2001-10-20 | | Release date: | 2002-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Complex between Pseudomonas Aeruginosa Alkaline Protease and its Cognate Inhibitor: Inhibition by a Zinc-NH2 Coordinative Bond
J.Biol.Chem., 276, 2001
|
|
1GO9
 
 | |
1GOA
 
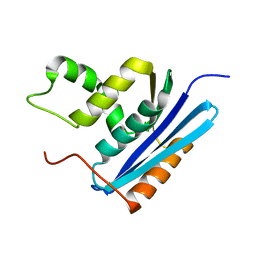 | | COOPERATIVE STABILIZATION OF ESCHERICHIA COLI RIBONUCLEASE HI BY INSERTION OF GLY-80B AND GLY-77-> ALA SUBSTITUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Nakamura, H, Morikawa, K, Kanaya, S. | | Deposit date: | 1993-05-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cooperative stabilization of Escherichia coli ribonuclease HI by insertion of Gly-80b and Gly-77-->Ala substitution.
Biochemistry, 32, 1993
|
|
