6Y33
 
 | | Streptavidin mutant S112R with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y2M
 
 | | Streptavidin mutant S112R with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y2T
 
 | | Streptavidin wildtype with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y3Q
 
 | | Streptavidin mutant S112R_K121E with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | SULFATE ION, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-18 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y25
 
 | | Streptavidin mutant S112R,K121E with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y34
 
 | | Streptavidin wildtype with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.307 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
4D6D
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6G
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6J
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6E
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6C
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98)(L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6F
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A, X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6I
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6H
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D71
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D72
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCOSIDE HYDROLASE, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
8YAX
 
 | |
8YB7
 
 | |
8YB5
 
 | |
8D1T
 
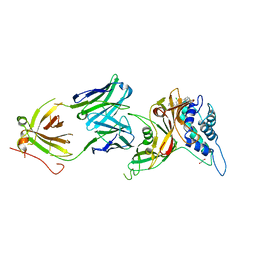 | | Crystal structure of human USP30 in complex with a covalent inhibitor 552 and a Fab | | Descriptor: | (1R,2R,4S,7E)-7-[amino(sulfanyl)methylidene]-2-{[(1P)-3-chloro-3'-(1-cyanocyclopropyl)[1,1'-biphenyl]-4-carbonyl]amino}-7-azabicyclo[2.2.1]heptan-7-ium, 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 30, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | TBD
To Be Published
|
|
8D0A
 
 | | Crystal structure of human USP30 in complex with a covalent inhibitor 829 and a Fab | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, mouse anti-huUSP30 Fab heavy chain, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | TBD
To Be Published
|
|
8F2E
 
 | |
5ARN
 
 | | Cu(I)-CSP3 (COPPER STORAGE PROTEIN 3) FROM METHYLOSINUS | | Descriptor: | COPPER (I) ION, CSP3 | | Authors: | Vita, N, Landolfi, G, Basle, A, Platsaki, S, Waldron, K, Dennison, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Cytosolic Copper Storage Proteins
To be Published
|
|
2JZD
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZE
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
