2G16
 
 | | Structure of S65A Y66S GFP variant after backbone fragmentation | | Descriptor: | Green fluorescent protein | | Authors: | Barondeau, D.P. | | Deposit date: | 2006-02-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding GFP Posttranslational Chemistry: Structures of Designed Variants that Achieve Backbone Fragmentation, Hydrolysis, and Decarboxylation.
J.Am.Chem.Soc., 128, 2006
|
|
3FB4
 
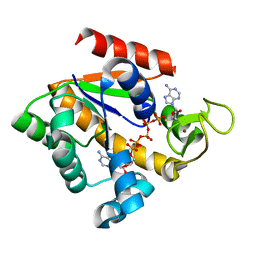 | | Crystal structure of adenylate kinase from Marinibacillus marinus | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Davlieva, M.G, Shamoo, Y. | | Deposit date: | 2008-11-18 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical characterization of an adenylate kinase originating from the psychrophilic organism Marinibacillus marinus.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1F3T
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE (ODC) COMPLEXED WITH PUTRESCINE, ODC'S REACTION PRODUCT. | | Descriptor: | 1,4-DIAMINOBUTANE, ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jackson, L.K, Brooks, H.B, Osterman, A.L, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Altering the reaction specificity of eukaryotic ornithine decarboxylase.
Biochemistry, 39, 2000
|
|
3HPR
 
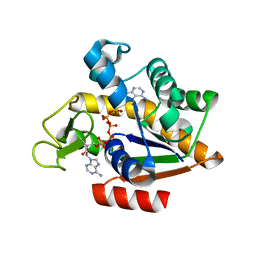 | |
2ZMW
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 6.0 | | Descriptor: | Fluorescent protein | | Authors: | Kikuchi, A, Fukumura, E, Karasawa, S, Mizuno, H, Miyawaki, A, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Thiazoline-Containing Chromophore in an Orange Fluorescent Protein, Monomeric Kusabira Orange
Biochemistry, 47, 2008
|
|
4UKD
 
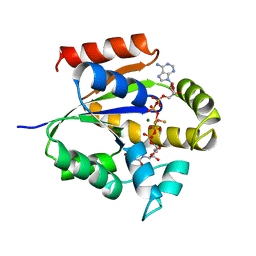 | | UMP/CMP KINASE FROM SLIME MOLD COMPLEXED WITH ADP, UDP, BERYLLIUM FLUORIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM DIFLUORIDE, MAGNESIUM ION, ... | | Authors: | Schlichting, I, Reinstein, J. | | Deposit date: | 1997-05-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
Biochemistry, 36, 1997
|
|
5X6I
 
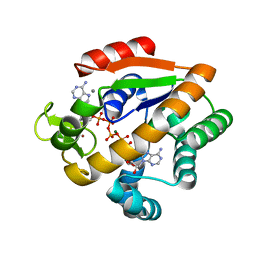 | | Crystal structure of B. subtilis adenylate kinase variant | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CALCIUM ION, ... | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mutational analyses of psychrophilic and mesophilic adenylate kinases highlight the role of hydrophobic interactions in protein thermal stability.
Struct Dyn., 6, 2019
|
|
2TOD
 
 | | ORNITHINE DECARBOXYLASE FROM TRYPANOSOMA BRUCEI K69A MUTANT IN COMPLEX WITH ALPHA-DIFLUOROMETHYLORNITHINE | | Descriptor: | ALPHA-DIFLUOROMETHYLORNITHINE, PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-05-18 | | Release date: | 1999-11-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
2G6X
 
 | |
2BTJ
 
 | | Fluorescent Protein EosFP - red form | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Photo-Induced Protein Cleavage and Green-to-Red Conversion of Fluorescent Protein Eosfp.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5AB5
 
 | |
1G7K
 
 | | CRYSTAL STRUCTURE OF DSRED, A RED FLUORESCENT PROTEIN FROM DISCOSOMA SP. RED | | Descriptor: | FLUORESCENT PROTEIN FP583 | | Authors: | Yarbrough, D, Wachter, R.M, Kallio, K, Matz, M.V, Remington, S.J. | | Deposit date: | 2000-11-10 | | Release date: | 2000-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2H8Q
 
 | |
3QAY
 
 | | Catalytic domain of CD27L endolysin targeting Clostridia Difficile | | Descriptor: | Endolysin, PHOSPHATE ION, ZINC ION | | Authors: | Mayer, M.J, Garefaliki, V, Spoerl, R, Narbad, A, Meijers, R. | | Deposit date: | 2011-01-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range.
J.Bacteriol., 193, 2011
|
|
3A28
 
 | | Crystal structure of L-2,3-butanediol dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, L-2.3-butanediol dehydrogenase, MAGNESIUM ION, ... | | Authors: | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | Deposit date: | 2009-05-02 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chiral substrate recognition by two 2,3-butanediol dehydrogenases
Febs Lett., 584, 2010
|
|
3EKH
 
 | | Calcium-saturated GCaMP2 T116V/K378W mutant monomer | | Descriptor: | CALCIUM ION, GLYCEROL, Myosin light chain kinase, ... | | Authors: | Akerboom, J, Velez Rivera, J.D, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the GCaMP Calcium Sensor Reveal the Mechanism of Fluorescence Signal Change and Aid Rational Design
J.Biol.Chem., 284, 2009
|
|
2Z6Y
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2UYC
 
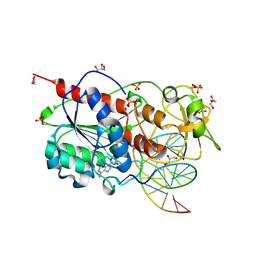 | |
1JC0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A REDUCED FORM | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, Aggeler, R, Oglesbee, D, Cannon, M, Capaldi, R.A, Tsien, R.Y, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
J.Biol.Chem., 279, 2004
|
|
2ARL
 
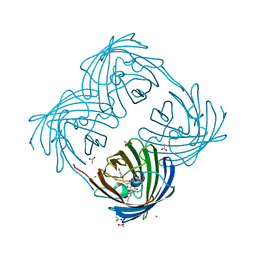 | | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding | | Descriptor: | ACETIC ACID, CHLORIDE ION, GFP-like non-fluorescent chromoprotein, ... | | Authors: | Wilmann, P.G, Battad, J, Beddoe, T, Olsen, S, Smith, S.C, Dove, S, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2005-08-19 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding
Photochem.Photobiol., 82, 2006
|
|
2VVI
 
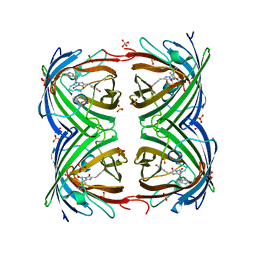 | | IrisFP fluorescent protein in its green form, trans conformation | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EVR
 
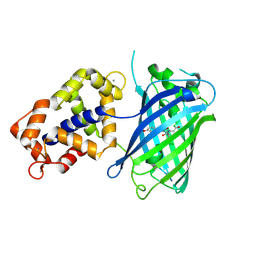 | | Crystal structure of Calcium bound monomeric GCAMP2 | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
1JS4
 
 | | ENDO/EXOCELLULASE:CELLOBIOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, ENDO/EXOCELLULASE E4, beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
2I3Y
 
 | | Crystal structure of human glutathione peroxidase 5 | | Descriptor: | 1,2-ETHANEDIOL, Epididymal secretory glutathione peroxidase | | Authors: | Kavanagh, K.L, Johansson, C, Rojkova, A, Umeano, C, Bunkoczi, G, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-21 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human glutathione peroxidase 5
To be published
|
|
2IB6
 
 | | Structural characterization of a blue chromoprotein and its yellow mutant from the sea anemone cnidopus japonicus | | Descriptor: | PHOSPHATE ION, Yellow mutant chromo protein | | Authors: | Chan, M.C.Y, Bosanac, I, Ho, D, Prive, G, Ikura, M. | | Deposit date: | 2006-09-10 | | Release date: | 2006-10-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Blue Chromoprotein and Its Yellow Mutant from the Sea Anemone Cnidopus Japonicus
J.Biol.Chem., 281, 2006
|
|
