1WYM
 
 | | Solution structure of the CH domain of human transgelin-2 | | Descriptor: | Transgelin-2 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-15 | | Release date: | 2005-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain of human transgelin-2
To be Published
|
|
1WF8
 
 | |
2DS4
 
 | | Solution structure of the filamin domain from human tripartite motif protein 45 | | Descriptor: | Tripartite motif protein 45 | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-21 | | Release date: | 2006-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the filamin domain from human tripartite motif protein 45
To be Published
|
|
2BWK
 
 | | Murine angiogenin, sulphate complex | | Descriptor: | ANGIOGENIN, SULFATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BWL
 
 | | Murine angiogenin, phosphate complex | | Descriptor: | ANGIOGENIN, PHOSPHATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BEF
 
 | | CRYSTAL STRUCTURE OF NDP KINASE COMPLEXED WITH MG, ADP, AND BEF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Xu, Y.W, Cherfils, J. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2KKW
 
 | | SLAS-micelle bound alpha-synuclein | | Descriptor: | Alpha-synuclein | | Authors: | Rao, J, Jao, C.C, Hegde, B, Langen, R, Ulmer, T.S. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | EPR, SOLUTION NMR | | Cite: | A combinatorial NMR and EPR approach for evaluating the structural ensemble of partially folded proteins.
J.Am.Chem.Soc., 132, 2010
|
|
2GCP
 
 | | Crystal structure of the human RhoC-GSP complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dias, S.M.G, Cerione, R.A. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystal Structures Reveal Two Activated States for RhoC.
Biochemistry, 46, 2007
|
|
2HW4
 
 | | Crystal structure of human phosphohistidine phosphatase | | Descriptor: | 14 kDa phosphohistidine phosphatase, FORMIC ACID | | Authors: | Busam, R.D, Thorsell, A.G, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Holmberg Schiavone, L, Hogbom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Stenmark, P, Sundstrom, M, Uppenberg, J, Van Den Berg, S, Weigelt, J, Persson, C, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-31 | | Release date: | 2006-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First structure of a eukaryotic phosphohistidine phosphatase
J.Biol.Chem., 281, 2006
|
|
2K76
 
 | | Solution structure of a paralog-specific Mena binder by NMR | | Descriptor: | pGolemi | | Authors: | Link, N.M, Hunke, C, Eichler, J, Bayer, P. | | Deposit date: | 2008-08-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of pGolemi, a high affinity Mena EVH1 binding miniature protein, suggests explanations for paralog-specific binding to Ena/VASP homology (EVH) 1 domains.
Biol.Chem., 390, 2009
|
|
2EOT
 
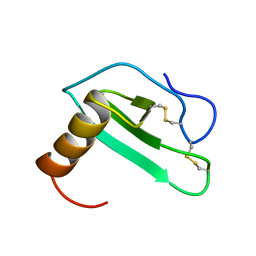 | | SOLUTION STRUCTURE OF EOTAXIN, AN ENSEMBLE OF 32 NMR SOLUTION STRUCTURES | | Descriptor: | EOTAXIN | | Authors: | Crump, M.P, Rajarathnam, K, Kim, K.-S, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1998-06-29 | | Release date: | 1998-11-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of eotaxin, a chemokine that selectively recruits eosinophils in allergic inflammation.
J.Biol.Chem., 273, 1998
|
|
2AI6
 
 | |
1ZFS
 
 | |
2A6A
 
 | |
2D9Q
 
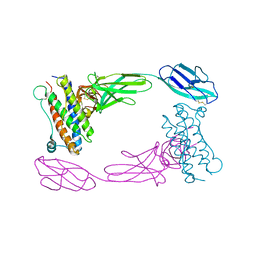 | | Crystal Structure of the Human GCSF-Receptor Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CSF3, Granulocyte colony-stimulating factor receptor | | Authors: | Tamada, T, Kuroki, R. | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1XAW
 
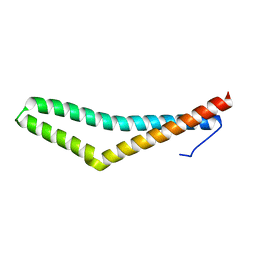 | |
1XLY
 
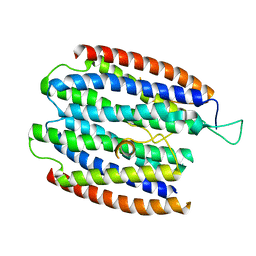 | | X-RAY STRUCTURE OF THE RNA-BINDING PROTEIN SHE2p | | Descriptor: | SHE2p | | Authors: | Niessing, D, Huettelmaier, S, Zenklusen, D, Singer, R.H, Burley, S.K. | | Deposit date: | 2004-09-30 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | She2p is a novel RNA binding protein with a basic helical hairpin motif
Cell(Cambridge,Mass.), 119, 2004
|
|
2XSZ
 
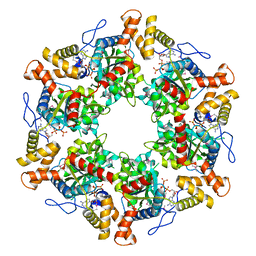 | | The dodecameric human RuvBL1:RuvBL2 complex with truncated domains II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RUVB-LIKE 1, RUVB-LIKE 2 | | Authors: | Gorynia, S, Bandeiras, T.M, Matias, P.M, Pinho, F.G, McVey, C.E, Vonrhein, C, Svergun, D.I, Round, A, Donner, P, Carrondo, M.A. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Insights Into a Dodecameric Molecular Machine - the Ruvbl1/Ruvbl2 Complex.
J.Struct.Biol., 176, 2011
|
|
6TXS
 
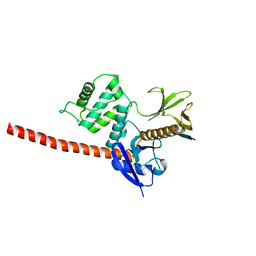 | | The structure of the FERM domain and helical linker of human moesin bound to a CD44 peptide | | Descriptor: | CD44 antigen, Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Kelly, J.J, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
7VDT
 
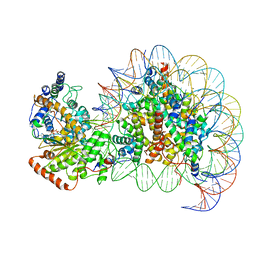 | | The motor-nucleosome module of human chromatin remodeling PBAF-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (207-MER), ... | | Authors: | Chen, Z.C, Chen, K.J, Yuan, J.J. | | Deposit date: | 2021-09-07 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of human chromatin-remodelling PBAF complex bound to a nucleosome.
Nature, 605, 2022
|
|
6TXQ
 
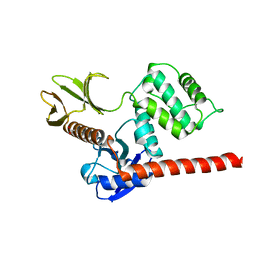 | | The high resolution structure of the FERM domain and helical linker of human moesin | | Descriptor: | ACETATE ION, Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Kelly, J.J, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
6U4K
 
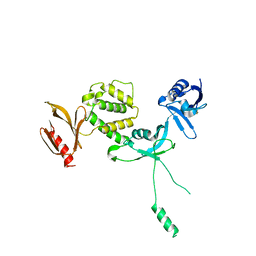 | | Human talin2 residues 1-403 | | Descriptor: | Talin-2 | | Authors: | Izard, T, Rangarajan, E.S, Colgan, L, Yasuda, R, Chinthalapudi, K. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (2.555 Å) | | Cite: | A distinct talin2 structure directs isoform specificity in cell adhesion.
J.Biol.Chem., 295, 2020
|
|
6V20
 
 | |
7TM3
 
 | | Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
7TUT
 
 | | Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-02-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
