4KT7
 
 | | The crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritolsynthase from Anaerococcus prevotii DSM 20548 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, CHLORIDE ION, SODIUM ION | | Authors: | Borek, D, Tan, K, Stols, L, Eschenfeidt, W.H, Otwinoski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-05 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritolsynthase from Anaerococcus prevotii DSM 20548
To be Published
|
|
3MU5
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2RK8
 
 | | The Structure of rat cytosolic PEPCK in complex with phosphonoformate | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Sullivan, S.M, Stiffin, R.M, Carlson, G.M, Holyoak, T. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Inhibition of Cytosolic PEPCK by Substrate Analogues. Kinetic and Structural Characterization of Inhibitor Recognition.
Biochemistry, 47, 2008
|
|
4L3A
 
 | |
3ENS
 
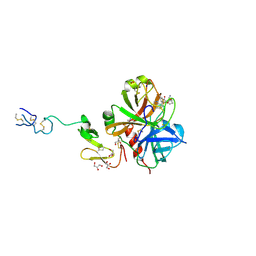 | |
4KXX
 
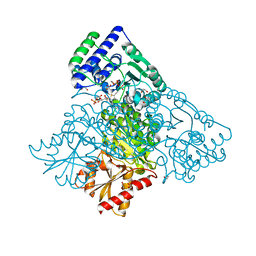 | | Human transketolase in covalent complex with donor ketose D-sedoheptulose-7-phosphate | | Descriptor: | (2R,3R,4S,5R,6S)-2,3,4,5,6,7-hexahydroxyheptyl dihydrogen phosphate, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
3ENZ
 
 | | Arsenolytic structure of Plasmodium falciparum purine nucleoside phosphorylase with hypoxanthine, ribose and arsenate ion | | Descriptor: | 1,4-anhydro-D-ribitol, ARSENATE, FORMIC ACID, ... | | Authors: | Chaikuad, A, Brady, R.L. | | Deposit date: | 2008-09-26 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases.
Bmc Struct.Biol., 9, 2009
|
|
3MTY
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. First step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4KXU
 
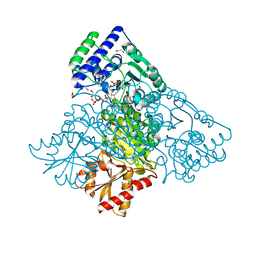 | | Human transketolase in covalent complex with donor ketose D-fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, D-SORBITOL-6-PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
3MU1
 
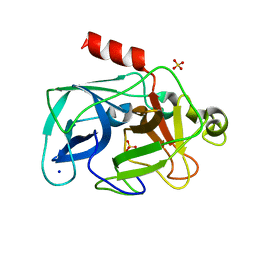 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4KZW
 
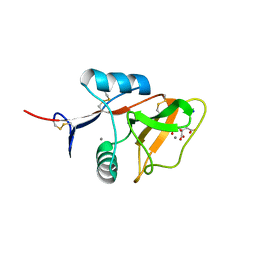 | | Structure of the carbohydrate-recognition domain of the C-type lectin mincle | | Descriptor: | C-TYPE LECTIN MINCLE, CALCIUM ION, CITRATE ANION, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rowntree, T.J.W, Guan, Y, Brash, M.A, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2013-05-30 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism for Recognition of an Unusual Mycobacterial Glycolipid by the Macrophage Receptor Mincle.
J.Biol.Chem., 288, 2013
|
|
4CNT
 
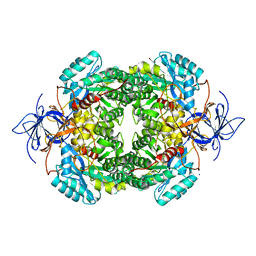 | | CRYSTAL STRUCTURE OF WT HUMAN CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-LIKE 3, SODIUM ION | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2RKA
 
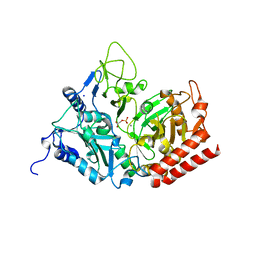 | | The Structure of rat cytosolic PEPCK in complex with phosphoglycolate | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, ... | | Authors: | Sullivan, S.M, Stiffin, R.M, Carlson, G.M, Holyoak, T. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential Inhibition of Cytosolic PEPCK by Substrate Analogues. Kinetic and Structural Characterization of Inhibitor Recognition.
Biochemistry, 47, 2008
|
|
4CT8
 
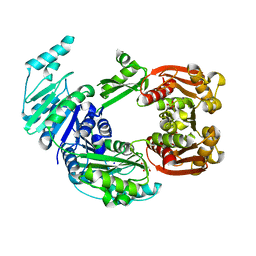 | |
3EB5
 
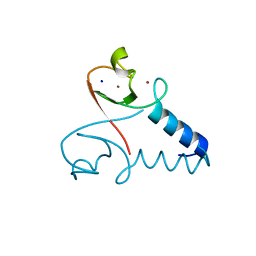 | | Structure of the cIAP2 RING domain | | Descriptor: | Baculoviral IAP repeat-containing protein 3, SODIUM ION, ZINC ION | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment.
J.Biol.Chem., 283, 2008
|
|
4MAT
 
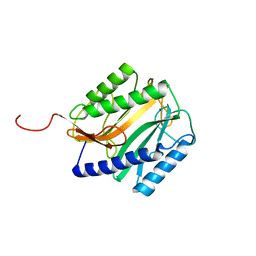 | | E.COLI METHIONINE AMINOPEPTIDASE HIS79ALA MUTANT | | Descriptor: | PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | Authors: | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | Deposit date: | 1999-03-29 | | Release date: | 1999-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
2DDB
 
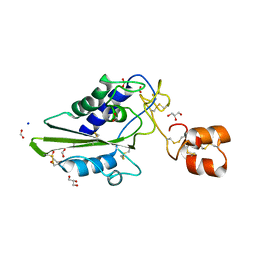 | | Crystal structure of pseudecin from Pseudechis porphyriacus | | Descriptor: | FORMIC ACID, GLYCEROL, Pseudecin, ... | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2006-01-25 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3EQ0
 
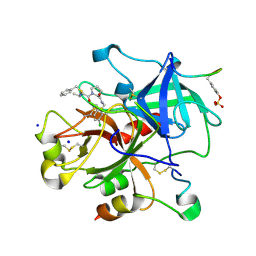 | | Thrombin Inhibitor | | Descriptor: | (2S)-N-[[2-(aminomethyl)-5-chloro-phenyl]methyl]-1-[(2R)-5-carbamimidamido-2-(phenylmethylsulfonylamino)pentanoyl]pyrrolidine-2-carboxamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G, Steinmetzer, T. | | Deposit date: | 2008-09-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Thrombin Inhibition
To be Published
|
|
3N79
 
 | | PduT C38S Mutant from Salmonella enterica Typhimurium | | Descriptor: | CHLORIDE ION, PduT, SODIUM ION, ... | | Authors: | Crowley, C.S, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insight into the Mechanisms of Transport across the Salmonella enterica Pdu Microcompartment Shell.
J.Biol.Chem., 285, 2010
|
|
7JYW
 
 | | Crystal Structure of HLA A*2402 in complex with TYQWIIRNW, an 9-mer influenza epitope | | Descriptor: | Beta-2-microglobulin, CALCIUM ION, MHC class I antigen, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C, Rossjohn, J. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
3ECQ
 
 | | Endo-alpha-N-acetylgalactosaminidase from Streptococcus pneumoniae: SeMet structure | | Descriptor: | CALCIUM ION, Endo-alpha-N-acetylgalactosaminidase, GLYCEROL, ... | | Authors: | Caines, M.E.C, Zhu, H, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structural basis for T-antigen hydrolysis by Streptococcus pneumoniae: a target for structure-based vaccine design.
J.Biol.Chem., 283, 2008
|
|
3EEB
 
 | | Structure of the V. cholerae RTX cysteine protease domain | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, SODIUM ION | | Authors: | Lupardus, P.J, Shen, A, Bogyo, M, Garcia, K.C. | | Deposit date: | 2008-09-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain
Science, 322, 2008
|
|
2RI0
 
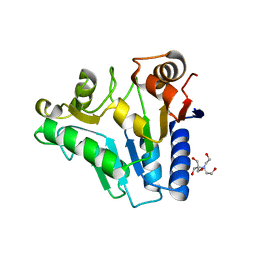 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucosamine-6-phosphate deaminase, SODIUM ION | | Authors: | Li, D, Liu, C, Li, L.F, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
4D2E
 
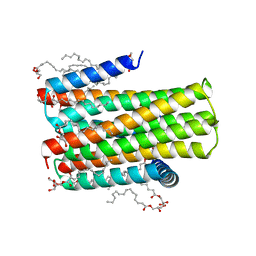 | | Crystal structure of an integral membrane kinase - v2.3 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, CITRATE ANION, ... | | Authors: | Li, D, Boland, C, Caffrey, M. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cell-Free Expression and in Meso Crystallisation of an Integral Membrane Kinase for Structure Determination.
Cell.Mol.Life Sci., 71, 2014
|
|
3NDT
 
 | | HIV-1 Protease Saquinavir:Ritonavir 1:1 complex structure | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2010-06-08 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Investigation of 2-Fold Disorder of Inhibitors and Relative Potency by Crystallizations of HIV-1 Protease in Ritonavir and Saquinavir Mixtures
Cryst.Growth Des., 11, 2011
|
|
