7B1G
 
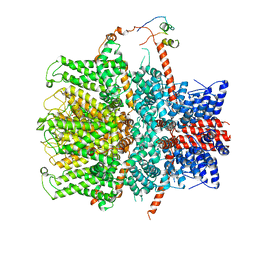 | | TRPC4 in complex with Calmodulin | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, CALCIUM ION, Calmodulin-1, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B1E
 
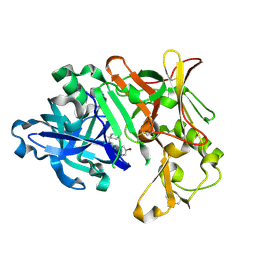 | | BACE1 IN COMPLEX WITH compound 3 (NB-641) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(4~{S})-2-azanyl-4-methyl-5,6-dihydro-1,3-thiazin-4-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KUE
 
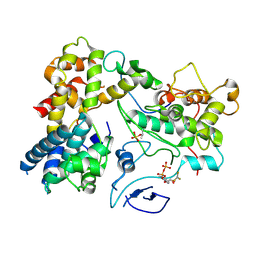 | | CryoEM structure of Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin CCL1, ... | | Authors: | van Eeuwen, T, Murakami, K, Li, T, Tsai, K.L. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of TFIIK for phosphorylation of CTD of RNA polymerase II.
Sci Adv, 7, 2021
|
|
7DKF
 
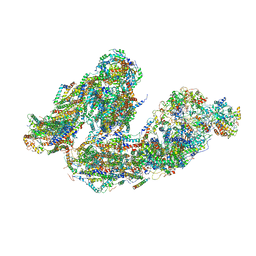 | | Activity optimized supercomplex state4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7B16
 
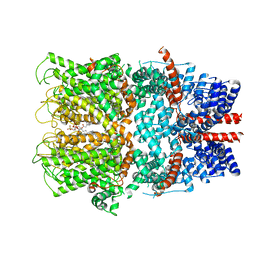 | | TRPC4 in complex with inhibitor GFB-9289 | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, 5-chloranyl-4-(4-cyclohexyl-3-oxidanylidene-piperazin-1-yl)-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B0Y
 
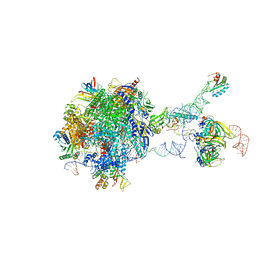 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
7KSK
 
 | |
7KSJ
 
 | |
7KSI
 
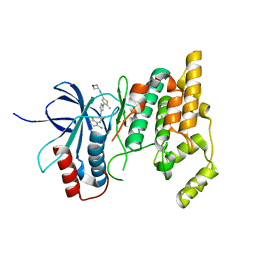 | |
7KSL
 
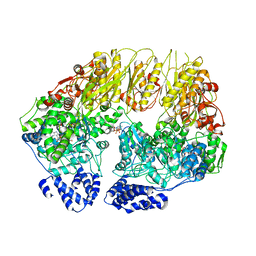 | | Substrate-free human mitochondrial LONP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7KSM
 
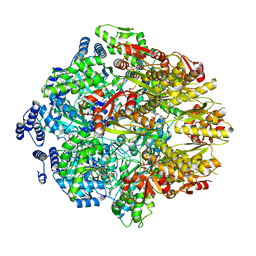 | | Human mitochondrial LONP1 with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7B0S
 
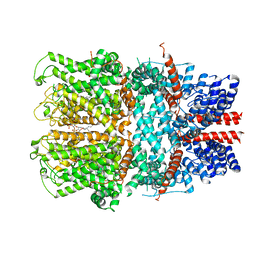 | | TRPC4 in complex with inhibitor GFB-8438 | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 5-chloranyl-4-[3-oxidanylidene-4-[[2-(trifluoromethyl)phenyl]methyl]piperazin-1-yl]-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7KSA
 
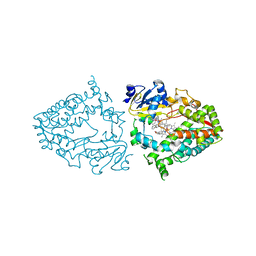 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (tert-butyl {1-[(1-oxo-3-phenyl-1-{[3-(pyridin-3-yl-kappaN)prop-1-en-1-yl]amino}propan-2-yl)sulfanyl]-3-phenylpropan-2-yl}carbamate)(6,6'-dimethyl-2,2'-bipyridine-kappa~2~N~1~,N~1'~)(1~2~,2~2~:2~6~,3~2~-terpyridine-kappa~3~N~1^{1~},N~2^{1~},N~3^{1~})ruthenium, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.S. | | Deposit date: | 2020-11-21 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Photosensitive Ru(II) Complexes as Inhibitors of the Major Human Drug Metabolizing Enzyme CYP3A4.
J.Am.Chem.Soc., 143, 2021
|
|
7KS8
 
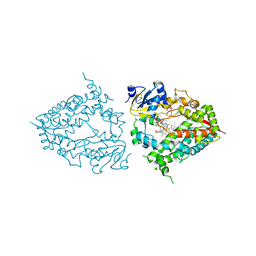 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (tert-butyl {1-[(3-oxo-3-{[(pyridin-3-yl-kappaN)methyl]amino}propyl)sulfanyl]-3-phenylpropan-2-yl}carbamate)(6,6'-dimethyl-2,2'-bipyridine-kappa~2~N~1~,N~1'~)(1~2~,2~2~:2~6~,3~2~-terpyridine-kappa~3~N~1^{1~},N~2^{1~},N~3^{1~})ruthenium, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I.S. | | Deposit date: | 2020-11-21 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Photosensitive Ru(II) Complexes as Inhibitors of the Major Human Drug Metabolizing Enzyme CYP3A4.
J.Am.Chem.Soc., 143, 2021
|
|
7KSE
 
 | | Crystal structure of Prototype Foamy Virus Protease-Reverse Transcriptase CSH mutant (selenomethionine-labeled) | | Descriptor: | CALCIUM ION, Peptidase A9/Reverse transcriptase/RNase H | | Authors: | Harrison, J.J.E.K, Das, K, Ruiz, F.X, Arnold, E. | | Deposit date: | 2020-11-21 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a Retroviral Polyprotein: Prototype Foamy Virus Protease-Reverse Transcriptase (PR-RT).
Viruses, 13, 2021
|
|
7KSF
 
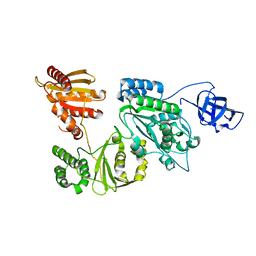 | | Crystal structure of Prototype Foamy Virus Protease-Reverse Transcriptase (native) | | Descriptor: | CALCIUM ION, Protease/Reverse transcriptase/ribonuclease H | | Authors: | Harrison, J.J.E.K, Das, K, Ruiz, F.X, Arnold, E. | | Deposit date: | 2020-11-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Retroviral Polyprotein: Prototype Foamy Virus Protease-Reverse Transcriptase (PR-RT).
Viruses, 13, 2021
|
|
7DJO
 
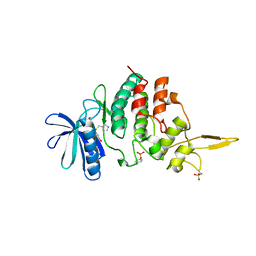 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 17 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-[[(3S)-pyrrolidin-3-yl]methylsulfanyl]acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7B0J
 
 | | TRPC4 in LMNG detergent | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, Transient receptor potential cation channel subfamily c member 4a | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7KRZ
 
 | | Human mitochondrial LONP1 in complex with Bortezomib | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Endogenous co-purified substrate, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-20 | | Release date: | 2021-02-24 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
