6VCY
 
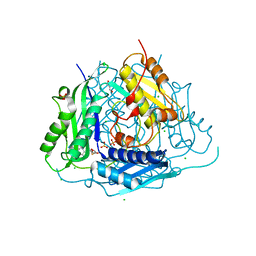 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 1 (AtMAT1) in complex with 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6VD2
 
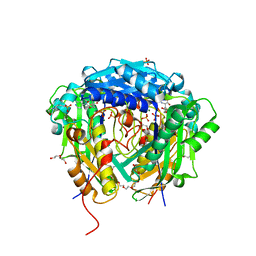 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with S-adenosylmethionine | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6VCX
 
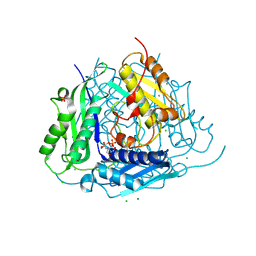 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 1 (AtMAT1) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6VD0
 
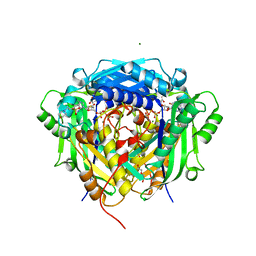 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with free Methionine and AMPCPP | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6VCZ
 
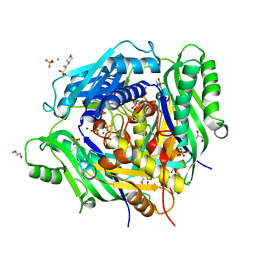 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) | | Descriptor: | 2-METHOXYETHANOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6VD1
 
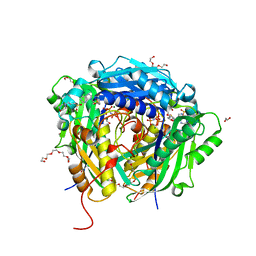 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with S-adenosylmethionine and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
7WIZ
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, GLUTAMINE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liu, J.L, Guo, C.J. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2OW3
 
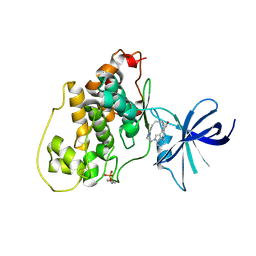 | | Glycogen synthase kinase-3 beta in complex with bis-(indole)maleimide pyridinophane inhibitor | | Descriptor: | BIS-(INDOLE)MALEIMIDE PYRIDINOPHANE, Glycogen synthase kinase-3 beta | | Authors: | Zhang, H.C, Bonaga, L.V, Ye, H, Derian, C.K, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2007-02-15 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel bis(indolyl)maleimide pyridinophanes that are potent, selective inhibitors of glycogen synthase kinase-3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3F88
 
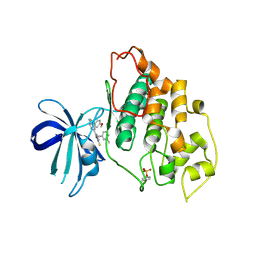 | | glycogen synthase Kinase 3beta inhibitor complex | | Descriptor: | 3-methylbenzonitrile, 5-[1-(4-methoxyphenyl)-1H-benzimidazol-6-yl]-1,3,4-oxadiazole-2(3H)-thione, Glycogen synthase kinase-3 beta | | Authors: | Mol, C.D, Dougan, D.R. | | Deposit date: | 2008-11-11 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and structure-activity relationships of 1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta.
Bioorg.Med.Chem., 17, 2009
|
|
5T31
 
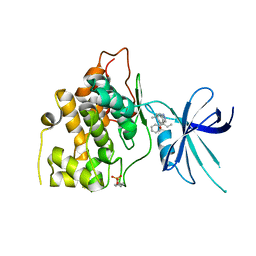 | | Exploiting an Asp-Glu switch in Glycogen Synthase Kinase 3 to design paralog selective inhibitors for use in acute myeloid leukemia | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Stein, A.J, Holson, E.B, Wagner, F.F, Cambell, A.J. | | Deposit date: | 2016-08-24 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
3F7Z
 
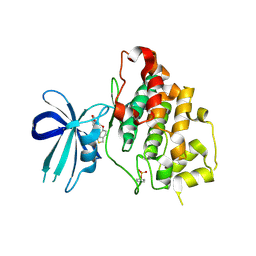 | | X-ray Co-Crystal Structure of Glycogen Synthase Kinase 3beta in Complex with an Inhibitor | | Descriptor: | 2-(1,3-benzodioxol-5-yl)-5-[(3-fluoro-4-methoxybenzyl)sulfanyl]-1,3,4-oxadiazole, Glycogen synthase kinase-3 beta | | Authors: | Mol, C.D, Dougan, D.R. | | Deposit date: | 2008-11-10 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, synthesis and structure-activity relationships of 1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta.
Bioorg.Med.Chem., 17, 2009
|
|
6HXO
 
 | |
7FTU
 
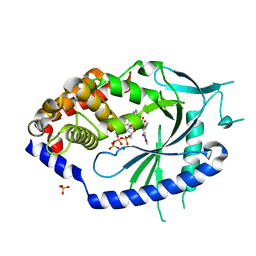 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with (Z)-N-[4-(4-chlorophenyl)sulfonylphenyl]-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide | | Descriptor: | (2Z)-N-[4-(4-chlorobenzene-1-sulfonyl)phenyl]-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide, ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
6B8J
 
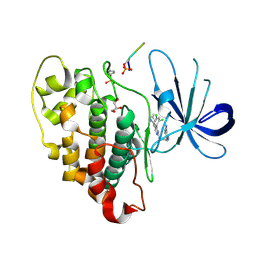 | |
1VDZ
 
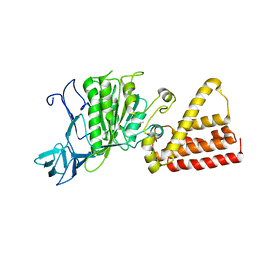 | | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, A-type ATPase subunit A | | Authors: | Maegawa, Y, Morita, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3
To be Published
|
|
1QDL
 
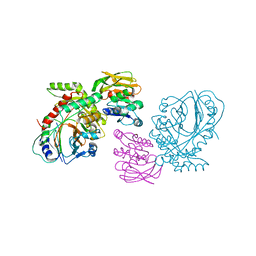 | | THE CRYSTAL STRUCTURE OF ANTHRANILATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | PROTEIN (ANTHRANILATE SYNTHASE (TRPE-SUBUNIT)), PROTEIN (ANTHRANILATE SYNTHASE (TRPG-SUBUNIT)) | | Authors: | Knoechel, T, Ivens, A, Hester, G, Gonzalez, A, Bauerle, R, Wilmanns, M, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1999-05-20 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7FTR
 
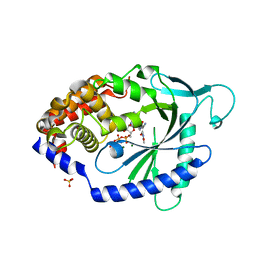 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with (Z)-N-(4-acetylphenyl)-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide | | Descriptor: | (2Z)-N-(4-acetylphenyl)-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide, ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
5TDZ
 
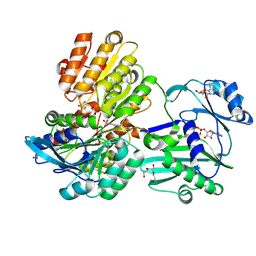 | |
5TES
 
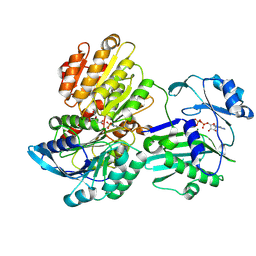 | |
5TDF
 
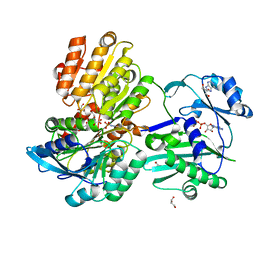 | |
5TDM
 
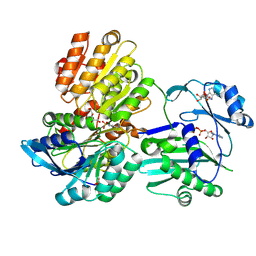 | |
2MX1
 
 | | Structure of the E. coli Threonylcarbamoyl-AMP Synthase TSAC | | Descriptor: | Threonylcarbamoyl-AMP synthase | | Authors: | Harris, K.A, Bobay, B.G, Sarachan, K.L, Sims, A.F, Bilbille, Y, Deutsch, C, Iwata-Reuyl, D, Agris, P.F. | | Deposit date: | 2014-12-06 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR-based Structural Analysis of Threonylcarbamoyl-AMP Synthase and Its Substrate Interactions.
J.Biol.Chem., 290, 2015
|
|
2Z02
 
 | | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase wit ATP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase from Methanocaldococcus jannaschii
To be Published
|
|
7FDA
 
 | | CryoEM Structure of Reconstituted V-ATPase, state1 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDC
 
 | | CryoEM Structures of Reconstituted V-ATPase, state3 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
