9FYI
 
 | |
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
9FYH
 
 | | Dye Type Peroxidase Aa from Streptomyces lividans by microcrystal electron diffraction (MicroED/3D ED) | | Descriptor: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hofer, G, Wang, L, Pacoste, L, Hager, P, Finjallaz, A, Williams, L, Worral, J, Steiner, R, Xu, H, Zou, X. | | Deposit date: | 2024-07-03 | | Release date: | 2025-07-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.4 Å) | | Cite: | Universal Serial electron diffraction for high quality protein structures
To Be Published
|
|
1MJD
 
 | | Structure of N-terminal domain of human doublecortin | | Descriptor: | DOUBLECORTIN | | Authors: | Kim, M.H, Cierpicki, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z.S. | | Deposit date: | 2002-08-27 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
9GP9
 
 | | Crystal Structure of Polyphosphate kinase 2-II (PPK2-II) from Lysinibacillus fusiformis bound to ADP (form I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Polyphosphate kinase | | Authors: | Saleem-Batcha, R, Keppler, M, Kuge, M, Andexer, J.N. | | Deposit date: | 2024-09-07 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights into Broad-Range Polyphosphate Kinase 2-II Enzymes Applicable for Pyrimidine Nucleoside Diphosphate Synthesis.
Chembiochem, 26, 2025
|
|
1MKZ
 
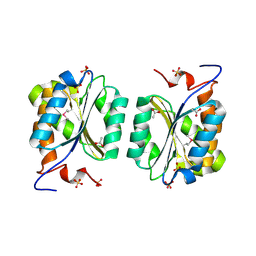 | | Crystal structure of MoaB protein at 1.6 A resolution. | | Descriptor: | ACETIC ACID, Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Sanishvili, R, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
J.Biol.Chem., 279, 2004
|
|
9FYS
 
 | | D11 mAbs bound to alpha-Bungarotoxin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-bungarotoxin, D11 mAbs Light chain, ... | | Authors: | Wade, J, Bohn, M.F, Laustsen, A.H, Morth, J.P. | | Deposit date: | 2024-07-03 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural mechanisms of a-neurotoxin-neutralizing antibodies enable rational design of pH-dependent antigen binding
To Be Published
|
|
1MBD
 
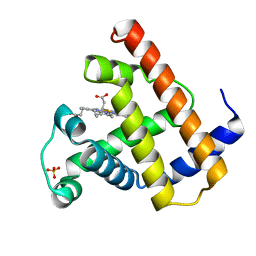 | |
1MBN
 
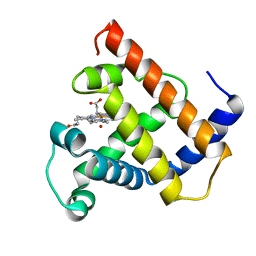 | |
9FXV
 
 | |
1MBV
 
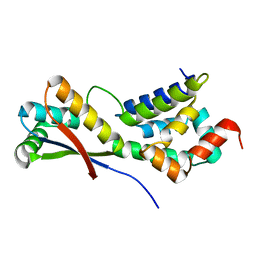 | | CRYSTAL STRUCTURE ANALYSIS OF ClpSN HETERODIMER TETRAGONAL FORM | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, Protein yljA | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of AAA+ Chaperone ClpA
J.Biol.Chem., 277, 2002
|
|
9FYX
 
 | | Influenza A/H7N9 polymerase pre-cleavage cap-snatching complex | | Descriptor: | 3' vRNA, 5'-vRNA, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-GUANOSINE, ... | | Authors: | Rotsch, A.H, Li, D, Dienemann, C, Cusack, S, Cramer, P. | | Deposit date: | 2024-07-04 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of Co-Transcriptional Cap-Snatching by Influenza Polymerase
To Be Published
|
|
1MC4
 
 | |
9G04
 
 | | Structure of MadB, a class I dehydrates from Clostridium maddingley in the apo state | | Descriptor: | Thiopeptide-type bacteriocin biosynthesis domain containing protein | | Authors: | Knospe, C.V, Ortiz, J, Reiners, J, Kedrov, A, Gertzen, C, Smits, S.H.J, Schmitt, L. | | Deposit date: | 2024-07-06 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the substrate binding mechanism of the class I dehydratase MadB.
Commun Biol, 8, 2025
|
|
1ML9
 
 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | Descriptor: | Histone H3 methyltransferase DIM-5, UNKNOWN LIGAND, ZINC ION | | Authors: | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | Deposit date: | 2002-08-30 | | Release date: | 2002-10-23 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
9G0J
 
 | | Structure of the PRO-PRO endopeptidase (PPEP-3) from Geobacillus thermodenitrificans | | Descriptor: | 1,2-ETHANEDIOL, ATLF-like domain-containing protein, BICINE, ... | | Authors: | Claushuis, B, Wojtalla, F, van Leeuwen, H, Corver, J, Baumann, U, Hensbergen, P. | | Deposit date: | 2024-07-08 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the PRO-PRO endopeptidase (PPEP-3) from Geobacillus thermodenitrificans
To Be Published
|
|
9GNX
 
 | | Human SENP5 in complex with SUMO1 (E67D) | | Descriptor: | Sentrin-specific protease 5, Small ubiquitin-related modifier 1 | | Authors: | Reverter, D, Sanchez-Alba, L, Maletic, M, Mulder, M. | | Deposit date: | 2024-09-04 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the human SENP5's SUMO isoform discrimination.
Nat Commun, 16, 2025
|
|
1MCE
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-B-ALA-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
7RXZ
 
 | | human Hsp90_MC domain structure | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-08-24 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | Structural basis of the key residue W320 responsible for Hsp90 conformational change.
J.Biomol.Struct.Dyn., 2022
|
|
7RY0
 
 | | human Hsp90_MC domain structure | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-08-24 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the key residue W320 responsible for Hsp90 conformational change.
J.Biomol.Struct.Dyn., 2022
|
|
1MCN
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-D-HIS-L-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
9FYT
 
 | | mAbs in complex with cobratoxin at pH 4.5 | | Descriptor: | Alpha-cobratoxin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wade, J, Bohn, M.F, Laustsen, A.H, Morth, J.P. | | Deposit date: | 2024-07-03 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural mechanisms of a-neurotoxin-neutralizing antibodies enable rational design of pH-dependent antigen binding
To Be Published
|
|
9G4T
 
 | |
1MCZ
 
 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA COMPLEXED WITH AN INHIBITOR, R-MANDELATE | | Descriptor: | (R)-MANDELIC ACID, BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Polovnikova, E.S, Bera, A.K, Hasson, M.S. | | Deposit date: | 2002-08-06 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Kinetic Analysis of Catalysis by a Thiamin
Diphosphate-Dependent Enzyme, Benzoylformate Decarboxylase
Biochemistry, 42, 2003
|
|
1MMF
 
 | | Crystal structure of substrate free form of glycerol dehydratase | | Descriptor: | COBALAMIN, POTASSIUM ION, glycerol dehydrase alpha subunit, ... | | Authors: | Liao, D.I, Dotson, G, Turner, I, Reiss, L, Emptage, M. | | Deposit date: | 2002-09-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of substrate free form of glycerol dehydratase
J.Inorg.Biochem., 93, 2003
|
|
