2PQ7
 
 | |
3IUZ
 
 | |
2ZWR
 
 | | Crystal structure of TTHA1623 from thermus thermophilus HB8 | | Descriptor: | Metallo-beta-lactamase superfamily protein, ZINC ION | | Authors: | Yamamura, A, Okada, A, Kameda, Y, Ohtsuka, J, Nakagawa, N, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of TTHA1623, a novel metallo-beta-lactamase superfamily protein from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3IJM
 
 | | The structure of a restriction endonuclease-like fold superfamily protein from Spirosoma linguale. | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Cuff, M.E, Tesar, C, Samano, S, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a restriction endonuclease-like fold superfamily protein from Spirosoma linguale.
TO BE PUBLISHED
|
|
3JQ1
 
 | |
3LEC
 
 | | The Crystal Structure of a protein in the NADB-Rossmann Superfamily from Streptococcus agalactiae to 1.8A | | Descriptor: | NADB-Rossmann Superfamily protein, SULFATE ION, ZINC ION | | Authors: | Stein, A.J, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of a protein in the NADB-Rossmann Superfamily from Streptococcus agalactiae to 1.8A
To be Published
|
|
2BKX
 
 | |
2BKV
 
 | |
4ORD
 
 | |
7AFS
 
 | | The structure of Artemis variant D37A | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AGI
 
 | | The structure of Artemis variant H35D | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7APV
 
 | | Structure of Artemis/DCLRE1C/SNM1C in complex with Ceftriaxone | | Descriptor: | 1,2-ETHANEDIOL, Ceftriaxone, NICKEL (II) ION, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
3MYV
 
 | |
3N1U
 
 | | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila | | Descriptor: | CALCIUM ION, Hydrolase, HAD superfamily, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-21 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila
To be published
|
|
3MCX
 
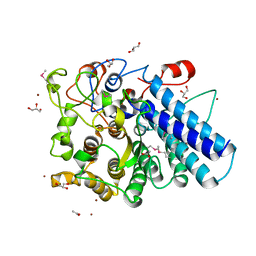 | |
2EQP
 
 | | Solution structure of the stn_TNFRSF12A_TNFR domain of Tumor necrosis factor receptor superfamily member 12A precursor | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the stn_TNFRSF12A_TNFR domain of Tumor necrosis factor receptor superfamily member 12A precursor
To be Published
|
|
2D11
 
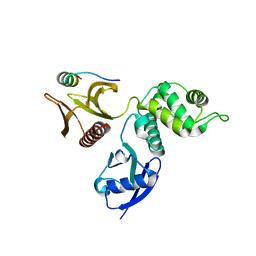 | |
2HX1
 
 | |
2DKC
 
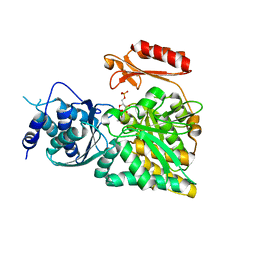 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the substrate complex | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
5DJB
 
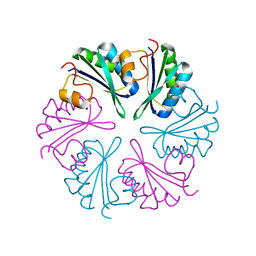 | |
5MP6
 
 | | Structure of the Unliganded Fab from HIV-1 Neutralizing Antibody CAP248-2B that Binds to the gp120 C-terminus - gp41 Interface, at two Angstrom resolution. | | Descriptor: | CAP248-2B Heavy Chain, CAP248-2B Light Chain, SULFATE ION | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structure and Recognition of a Novel HIV-1 gp120-gp41 Interface Antibody that Caused MPER Exposure through Viral Escape.
PLoS Pathog., 13, 2017
|
|
2DKD
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the product complex | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-galactopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
4OAE
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 C29A/C117A/Y128A mutant in complex with chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, GNAT superfamily acetyltransferase PA4794, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of a GNAT superfamily acetyltransferase PA4794 C29A/C117A/Y128A mutant in complex with chloramphenicol
To be Published
|
|
2J0V
 
 | | The crystal structure of Arabidopsis thaliana RAC7-ROP9: the first RAS superfamily GTPase from the plant kingdom | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAC-LIKE GTP-BINDING PROTEIN ARAC7 | | Authors: | Sormo, C.G, Leiros, I, Brembu, T, Winge, P, Os, V, Bones, A.M. | | Deposit date: | 2006-08-07 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Crystal Structure of Arabidopsis Thaliana Rac7/Rop9: The First Ras Superfamily Gtpase from the Plant Kingdom.
Phytochemistry, 67, 2006
|
|
6TT5
 
 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
