1FGQ
 
 | |
1FGR
 
 | |
1FGS
 
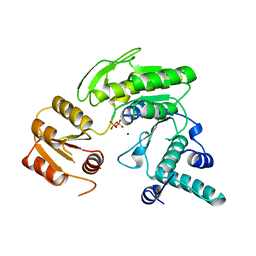 | | FOLYLPOLYGLUTAMATE SYNTHETASE FROM LACTOBACILLUS CASEI | | Descriptor: | FOLYLPOLYGLUTAMATE SYNTHETASE, MAGNESIUM ION, PYROPHOSPHATE 2- | | Authors: | Sun, X, Bognar, A, Baker, E, Smith, C. | | Deposit date: | 1998-04-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural homologies with ATP- and folate-binding enzymes in the crystal structure of folylpolyglutamate synthetase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1FGT
 
 | |
1FGU
 
 | |
1FGV
 
 | |
1FGX
 
 | | CRYSTAL STRUCTURE OF THE BOVINE BETA 1,4 GALACTOSYLTRANSFERASE (B4GALT1) CATALYTIC DOMAIN COMPLEXED WITH UMP | | Descriptor: | BETA 1,4 GALACTOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Gastinel, L.N, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-07-29 | | Release date: | 2000-08-16 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the bovine beta4galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose.
EMBO J., 18, 1999
|
|
1FGY
 
 | | GRP1 PH DOMAIN WITH INS(1,3,4,5)P4 | | Descriptor: | GRP1, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Lietzke, S.E, Bose, S, Cronin, T, Klarlund, J, Chawla, A, Czech, M.P, Lambright, D.G. | | Deposit date: | 2000-07-29 | | Release date: | 2000-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
1FGZ
 
 | | GRP1 PH DOMAIN (UNLIGANDED) | | Descriptor: | GRP1, SULFATE ION | | Authors: | Lietzke, S.E, Bose, S, Cronin, T, Klarlund, J, Chawla, A, Czech, M.P, Lambright, D.G. | | Deposit date: | 2000-07-29 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
1FH0
 
 | |
1FH1
 
 | |
1FH2
 
 | |
1FH3
 
 | |
1FH5
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MONOCLONAL ANTIBODY MAK33 | | Descriptor: | MONOCLONAL ANTIBODY MAK33 | | Authors: | Augustine, J.G, de la Calle, A, Knarr, G, Buchner, J, Frederick, C.A. | | Deposit date: | 2000-07-31 | | Release date: | 2000-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the fab fragment of the monoclonal antibody MAK33. Implications for folding and interaction with the chaperone bip.
J.Biol.Chem., 276, 2001
|
|
1FH7
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED INHIBITOR DEOXYNOJIRIMYCIN | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FH8
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED ISOFAGOMINE INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FH9
 
 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED LACTAM OXIME INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-(2Z,3S,4S,5R)-2-hydroxyiminopiperidine-3,4,5-triol | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FHA
 
 | |
1FHB
 
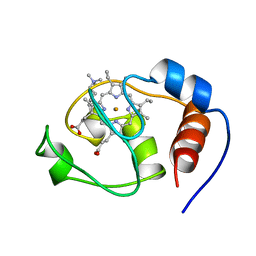 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE CYANIDE ADDUCT OF A MET80ALA VARIANT OF SACCHAROMYCES CEREVISIAE ISO-1-CYTOCHROME C. IDENTIFICATION OF LIGAND-RESIDUE INTERACTIONS IN THE DISTAL HEME CAVITY | | Descriptor: | CYANIDE ION, FERRICYTOCHROME C, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Bren, K.L, Gray, H.B, Sompornpisut, P, Turano, P. | | Deposit date: | 1995-06-16 | | Release date: | 1995-09-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Solution Structure of the Cyanide Adduct of a met80Ala Variant of Saccharomyces Cerevisiae Iso-1-Cytochrome C. Identification of Ligand-Residue Interactions in the Distal Heme Cavity
Biochemistry, 34, 1995
|
|
1FHD
 
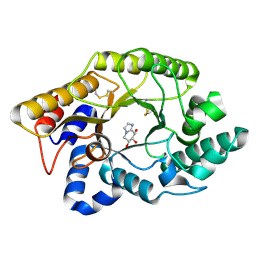 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED IMIDAZOLE INHIBITOR | | Descriptor: | 5,6,7,8-TETRAHYDRO-IMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, BETA-1,4-XYLANASE, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FHE
 
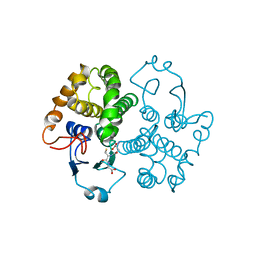 | | GLUTATHIONE TRANSFERASE (FH47) FROM FASCIOLA HEPATICA | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization, structural determination and analysis of a novel parasite vaccine candidate: Fasciola hepatica glutathione S-transferase.
J.Mol.Biol., 273, 1997
|
|
1FHF
 
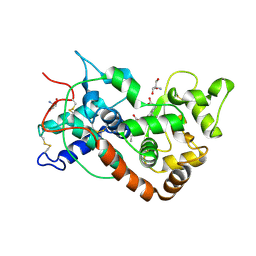 | | THE STRUCTURE OF SOYBEAN PEROXIDASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Henriksen, A, Mirza, O, Indiana, C, Welinder, K, Teilum, K, Gajhede, M. | | Deposit date: | 2000-08-01 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of soybean seed coat peroxidase: a plant peroxidase with unusual stability and haem-apoprotein interactions.
Protein Sci., 10, 2001
|
|
1FHG
 
 | | HIGH RESOLUTION REFINEMENT OF TELOKIN | | Descriptor: | TELOKIN | | Authors: | Tomchick, D.R, Minor, W, Kiyatkin, A, Lewinski, K, Somlyo, A.V, Somlyo, A.P. | | Deposit date: | 2000-08-01 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
1FHH
 
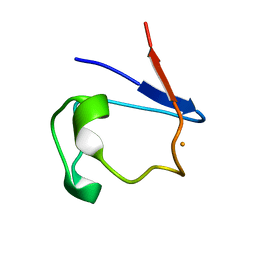 | | X-RAY CRYSTAL STRUCTURE OF OXIDIZED RUBREDOXIN | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Min, T, Ergenekan, C.E, Eidsness, M.K, Ichiye, T, Kang, C. | | Deposit date: | 2000-08-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Leucine 41 is a gate for water entry in the reduction of Clostridium pasteurianum rubredoxin.
Protein Sci., 10, 2001
|
|
1FHI
 
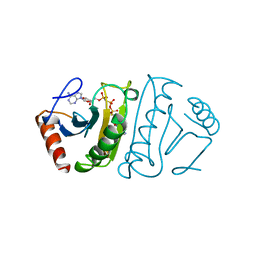 | | SUBSTRATE ANALOG (IB2) COMPLEX WITH THE FRAGILE HISTIDINE TRIAD PROTEIN, FHIT | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN, P1-P2-METHYLENE-P3-THIO-DIADENOSINE TRIPHOSPHATE | | Authors: | Pace, H.C, Garrison, P.N, Barnes, L.D, Draganescu, A, Rosler, A, Blackburn, G.M, Siprashvili, Z, Croce, C.M, Huebner, K, Brenner, C. | | Deposit date: | 1997-12-11 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Genetic, biochemical, and crystallographic characterization of Fhit-substrate complexes as the active signaling form of Fhit.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
