9F5F
 
 | | UP1 in complex with Z992569480 | | Descriptor: | (3R)-N-methyl-1-(pyridazin-3-yl)piperidin-3-amine, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4D
 
 | | UP1 in complex with Z1203107138 | | Descriptor: | 1-ethyl-N-[(4-fluorophenyl)methyl]-1H-pyrazole-4-carboxamide, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-27 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4N
 
 | | UP1 in complex with Z137811222 | | Descriptor: | 1-[4-(3-phenylpropyl)piperazin-1-yl]ethan-1-one, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4G
 
 | | UP1 in complex with Z1373445602 | | Descriptor: | 4-(3-fluoranylpyridin-2-yl)-1-methyl-piperazin-2-one, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-27 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4U
 
 | | UP1 in complex with Z111529496 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N}-(1~{H}-benzimidazol-2-ylmethyl)-2-methoxy-ethanamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4P
 
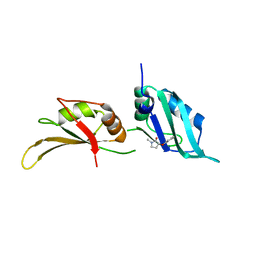 | | UP1 in complex with Z1217960891 | | Descriptor: | (3S)-3-(3-fluorophenoxy)-1-methylpyrrolidin-2-one, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F54
 
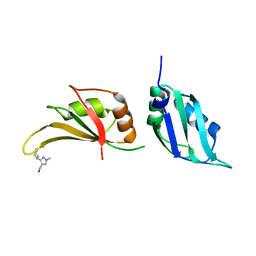 | | UP1 in complex with EN300-197154 | | Descriptor: | 1-methyl-3-[(3R)-piperidin-3-yl]-1H-pyrazole-4-carboxamide, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F50
 
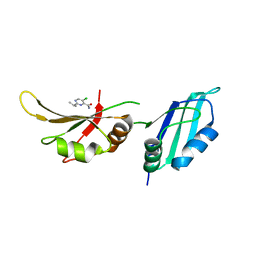 | | UP1 in complex with Z906021418 | | Descriptor: | 5-chloro-2-(propan-2-yl)pyrimidine-4-carboxamide, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4H
 
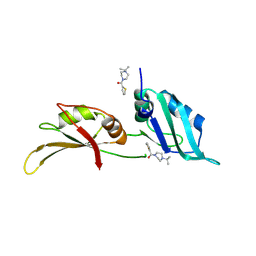 | | UP1 in complex with Z106579662 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, [4-(propan-2-yl)piperazin-1-yl](thiophen-2-yl)methanone | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-27 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F5K
 
 | | UP1 in complex with Z33546965 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N}-(2-azanyl-2-oxidanylidene-ethyl)-4-methoxy-benzamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4T
 
 | | UP1 in complex with EN300-805013 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, N-(4-methoxyphenyl)glycinamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4S
 
 | | UP1 in complex with Z56880342 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4Y
 
 | | UP1 in complex with Z90120418 | | Descriptor: | 1-(3,4,5-trimethoxyphenyl)methanamine, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4Q
 
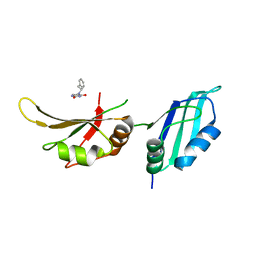 | | UP1 in complex with Z641239276 | | Descriptor: | 5-(2-phenylethylamino)pyrimidine-2,4-diol, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F51
 
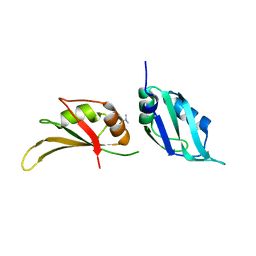 | | UP1 in complex with Z608065044 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N},~{N}-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4F
 
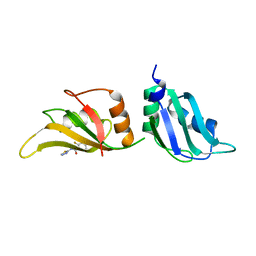 | | UP1 in complex with Z86417414 | | Descriptor: | 2-fluoranyl-~{N}-(1,3,4-thiadiazol-2-yl)benzamide, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-27 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F5D
 
 | | UP1 in complex with Z641230552 | | Descriptor: | 5-[(phenylmethyl)amino]-1~{H}-pyrimidine-2,4-dione, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4O
 
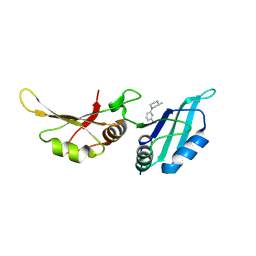 | | UP1 in complex with Z991506900 | | Descriptor: | (3~{R})-3-methyl-1-(6-methylpyridin-2-yl)piperazine, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F52
 
 | | UP1 in complex with Z734147462 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, [4-(methylamino)oxan-4-yl]methanol | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4K
 
 | | UP1 in complex with ZINC72259689 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N}-(cyclobutylmethyl)-1,3-dimethyl-pyrazole-4-carboxamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F55
 
 | | UP1 in complex with Z235361315 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N}-(3-azanyl-2-methyl-phenyl)propanamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
3QX3
 
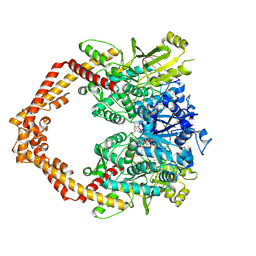 | | Human topoisomerase IIbeta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wu, C.C, Li, T.K, Farh, L, Lin, L.Y, Lin, T.S, Yu, Y.J, Yen, T.J, Chiang, C.W, Chan, N.L. | | Deposit date: | 2011-03-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide
Science, 333, 2011
|
|
3QZS
 
 | |
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | Deposit date: | 2003-12-15 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
9F4R
 
 | | UP1 in complex with Z802821712 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, N-ethyl-6-methylpyridazin-3-amine | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
