7MH3
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-chlorotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J.B, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MHA
 
 | |
2D2C
 
 | | Crystal Structure Of Cytochrome B6F Complex with DBMIB From M. Laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, Apocytochrome f, ... | | Authors: | Yan, J, Kurisu, G, Cramer, W.A. | | Deposit date: | 2005-09-07 | | Release date: | 2005-12-13 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7EQD
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOSPIRILLUM RUBRUM | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-azanyl-5-[(2~{E},6~{E},8~{E},10~{E},12~{E},14~{E},18~{E},22~{E},26~{E},30~{E},34~{E})-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,8,10,12,14,18,22,26,30,34,38-dodecaenyl]-3-methoxy-6-methyl-cyclohexa-2,5-diene-1,4-dione, CARDIOLIPIN, ... | | Authors: | Tani, K, Kanno, R, Ji, X.-C, Yu, L.-J, Hall, M, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-05-01 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM Structure of the Photosynthetic LH1-RC Complex from Rhodospirillum rubrum .
Biochemistry, 2021
|
|
5Y5S
 
 | | Structure of photosynthetic LH1-RC super-complex at 1.9 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Yu, L.-J, Suga, M, Wang-Otomo, Z.-Y, Shen, J.-R. | | Deposit date: | 2017-08-09 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of photosynthetic LH1-RC supercomplex at 1.9 angstrom resolution.
Nature, 556, 2018
|
|
7F0L
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES MONOMER | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna pigment protein beta chain, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tani, K, Nagashima, V.P, Kanno, R, Kawamura, S, Kikuchi, R, Ji, X.-C, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-10 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | A previously unrecognized membrane protein in the Rhodobacter sphaeroides LH1-RC photocomplex.
Nat Commun, 12, 2021
|
|
5YQ7
 
 | | Cryo-EM structure of the RC-LH core complex from Roseiflexus castenholzii | | Descriptor: | 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, BACTERIOCHLOROPHYLL A, ... | | Authors: | Shi, Y, Xin, Y.Y, Niu, T.X, Wang, Q.Q, Niu, W.Q, Huang, X.J, Ding, W, Blankenship, R.E, Xu, X.L, Sun, F. | | Deposit date: | 2017-11-05 | | Release date: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote.
Nat Commun, 9, 2018
|
|
7C9R
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF THIORHODOVIBRIO STRAIN 970 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26-undecaene, Alpha subunit 1 of light-harvesting 1 complex, ... | | Authors: | Tani, K, Kanno, R, Makino, Y, Hall, M, Takenouchi, M, Imanishi, M, Yu, L.-J, Overmann, J, Madigan, M.T, Kimura, Y, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2020-06-07 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of a Ca 2+ -bound photosynthetic LH1-RC complex containing multiple alpha beta-polypeptides.
Nat Commun, 11, 2020
|
|
7C52
 
 | | Co-crystal structure of a photosynthetic LH1-RC in complex with electron donor HiPIP | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Yu, L.-J, Wang-Otomo, Z.-Y. | | Deposit date: | 2020-05-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of a photosynthetic LH1-RC in complex with its electron donor HiPIP.
Nat Commun, 12, 2021
|
|
8FUO
 
 | | Fe-bound AibH1H2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, FE (III) ION | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
5LSE
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH Glu L212 replaced with Ala (CHAIN L, EL212W), Asp L213 replaced with ALA (Chain L, DL213A) AND LEU M215 REPLACED WITH ALA (CHAIN M, LM215A) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Jones, M.R. | | Deposit date: | 2016-08-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | On the mechanism of ubiquinone mediated photocurrent generation by a reaction center based photocathode.
Biochim.Biophys.Acta, 1857, 2016
|
|
5LRI
 
 | |
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
8FUL
 
 | |
8FUM
 
 | | AibH1H2 metalated with Fe in the presence of Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
8HJV
 
 | | Cryo-EM structure of carotenoid-depleted RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8HJU
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
2MQH
 
 | |
1PRC
 
 | | CRYSTALLOGRAPHIC REFINEMENT AT 2.3 ANGSTROMS RESOLUTION AND REFINED MODEL OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS | | Descriptor: | 15-trans-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Deisenhofer, J, Epp, O, Miki, K, Huber, R, Michel, H. | | Deposit date: | 1988-02-04 | | Release date: | 1989-01-09 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic refinement at 2.3 A resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis.
J.Mol.Biol., 246, 1995
|
|
1PCR
 
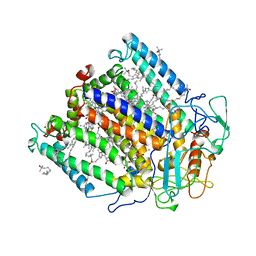 | | STRUCTURE OF THE PHOTOSYNTHETIC REACTION CENTRE FROM RHODOBACTER SPHAEROIDES AT 2.65 ANGSTROMS RESOLUTION: COFACTORS AND PROTEIN-COFACTOR INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Ermler, U, Fritzsch, G, Michel, H. | | Deposit date: | 1994-11-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 A resolution: cofactors and protein-cofactor interactions.
Structure, 2, 1994
|
|
1PSS
 
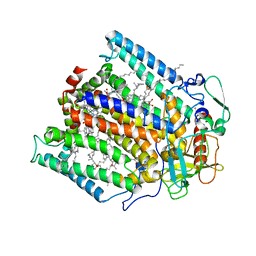 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
1PST
 
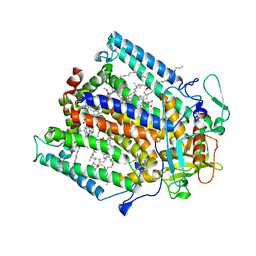 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
2RCR
 
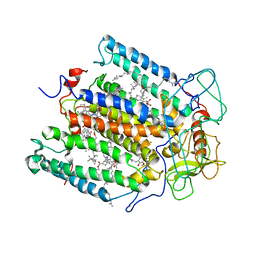 | | STRUCTURE OF THE MEMBRANE-BOUND PROTEIN PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, Coenzyme Q10, ... | | Authors: | Chang, C.-H, Norris, J, Schiffer, M. | | Deposit date: | 1991-02-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the membrane-bound protein photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 30, 1991
|
|
2JBL
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM BLASTOCHLORIS VIRIDIS | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2006-12-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Comparison of Stigmatellin Conformations, Free and Bound to the Photosynthetic Reaction Center and the Cytochrome Bc(1) Complex.
J.Mol.Biol., 368, 2007
|
|
4IC6
 
 | | Crystal structure of Deg8 | | Descriptor: | Protease Do-like 8, chloroplastic | | Authors: | Gong, W, Sun, W, Fan, H, Gao, F, Liu, L. | | Deposit date: | 2012-12-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
