5SPJ
 
 | |
5SEZ
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cc(nc(c1)CC)Cl)C(Nc3cc2nc(nn2cc3)c4ccccc4)=O, micromolar IC50=0.010134 | | Descriptor: | 2-chloro-6-ethyl-N-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]pyridine-4-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SF5
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1cc(nn2c1nc(c2C)C)CCc3nc(cn3C)c4ccccc4, micromolar IC50=0.0034475 | | Descriptor: | (4S)-2,3-dimethyl-6-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl]imidazo[1,2-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFF
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1cc(nn2c1nc(c2CO)C)C#Cc3nc(cn3C)c4ccccc4, micromolar IC50=0.00103 | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SPM
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with FRESH00002410346 | | Descriptor: | 4-hydroxy-6-(3-hydroxy-1-methyl-1,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridine-6-carbonyl)-2H-pyran-2-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3KXM
 
 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor K74 | | Descriptor: | Casein kinase II subunit alpha, N-methyl-2-[(4,5,6,7-tetrabromo-1-methyl-1H-benzimidazol-2-yl)sulfanyl]acetamide | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
2W7A
 
 | |
5UR9
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP5 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, epidermal, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19800353 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
4QN9
 
 | | Structure of human NAPE-PLD | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 1,2-Distearoyl-sn-glycerophosphoethanolamine, N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D, ... | | Authors: | Garau, G. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Structure of human N-acylphosphatidylethanolamine-hydrolyzing phospholipase D: regulation of fatty acid ethanolamide biosynthesis by bile acids.
Structure, 23, 2015
|
|
5ZH4
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-7 | | Descriptor: | (3R)-6,8-dihydroxy-3-{[(2S,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
5UOX
 
 | |
7KEU
 
 | | Cryo-EM structure of the Caspase-1-CARD:ASC-CARD octamer | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD, Caspase-1 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Ruan, J, Wu, H. | | Deposit date: | 2020-10-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
5V0X
 
 | |
1KTW
 
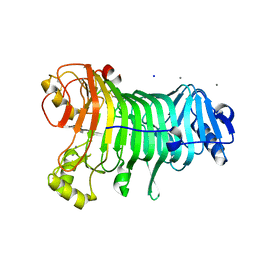 | | IOTA-CARRAGEENASE COMPLEXED TO IOTA-CARRAGEENAN FRAGMENTS | | Descriptor: | 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Michel, G, Kahn, R, Dideberg, O. | | Deposit date: | 2002-01-18 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Bases of the Processive Degradation of iota-Carrageenan, a Main Cell Wall Polysaccharide of Red Algae.
J.Mol.Biol., 334, 2003
|
|
3GTC
 
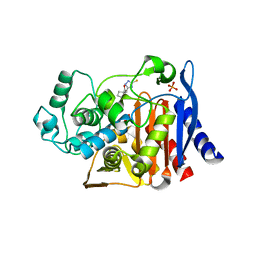 | | AmpC beta-lactamase in complex with Fragment-based Inhibitor | | Descriptor: | (1R,2S)-2-(5-thioxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)cyclohexanecarboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Teotico, D.T, Shoichet, B.K. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Docking for fragment inhibitors of AmpC beta-lactamase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7DVK
 
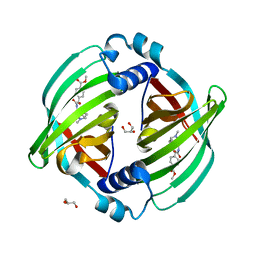 | | PyrI4 in complex with intermolecular Diels-Alder product | | Descriptor: | GLYCEROL, Spiro-conjugate synthase, methyl 4-[[(1S,6S)-6-(dimethylcarbamoyl)cyclohex-2-en-1-yl]carbamoyloxymethyl]benzoate | | Authors: | Kashyap, R, Addlagatta, A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exo-selective intermolecular Diels-Alder reaction by PyrI4 and AbnU on non-natural substrates.
Commun Chem, 2021
|
|
3I85
 
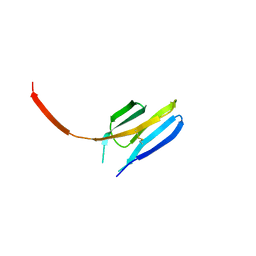 | |
4CC5
 
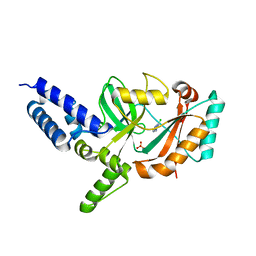 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
3IDO
 
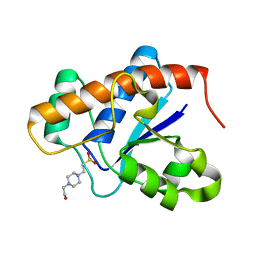 | |
4C1A
 
 | |
4TYH
 
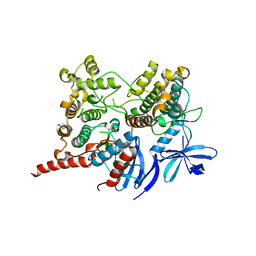 | | Ternary complex of P38 and MK2 with a P38 inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14, N-[5-(dimethylsulfamoyl)-2-methylphenyl]-1-phenyl-5-propyl-1H-pyrazole-4-carboxamide | | Authors: | Cumming, J.G, Debreczeni, J.E, Edfeldt, F, Evertsson, F, Harrison, M, Holdgate, G, James, M, Lamont, S, Oldham, K, Sullivan, J.E, Wells, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of substrate selective, ATP-competitive P38 alpha MAP kinase inhibitors
To Be Published
|
|
5UMC
 
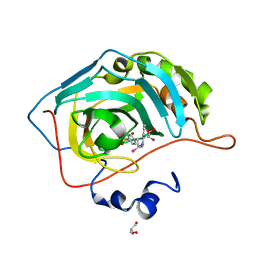 | | Synthesis of novel seleno ureido containing compounds as SLC-0111 analogs. Investigations on carbonic anhydrases activity, glutathione peroxidase and X-ray crystallography | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, UNKNOWN LIGAND, ... | | Authors: | Peat, T.S, Angeli, A, Tanini, D, Bartolucci, G, Capperucci, A, Supuran, C.T, Carta, F. | | Deposit date: | 2017-01-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
3HBC
 
 | | Crystal Structure of Choloylglycine Hydrolase from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, Choloylglycine hydrolase, GLYCEROL | | Authors: | Kim, Y, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Crystal Structure of Choloylglycine Hydrolase from Bacteroides thetaiotaomicron VPI
To be Published
|
|
4TZD
 
 | | Crystal structure of Canavalia maritima lectin (ConM) complexed with interleukin - 1 beta primer | | Descriptor: | Concanavalin-A, DNA (5'-D(P*CP*G)-3'), DNA (5'-D(P*TP*C)-3') | | Authors: | Vieira, D.B.H.A, Delatorre, P, Rocha, B.A.M, Teixeira, C.S, Silva-Filho, J.C, Lima, E.M, Nobrega, R.B, Cavada, B.S. | | Deposit date: | 2014-07-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Canavalia maritima lectin (ConM) complexed with interleukin 1 - beta primer
To Be Published
|
|
4C6A
 
 | | High Resolution Structure of the Nucleoside diphosphate kinase | | Descriptor: | DI(HYDROXYETHYL)ETHER, NUCLEOSIDE DIPHOSPHATE KINASE, CYTOSOLIC | | Authors: | Priet, S, Ferron, F, Alvarez, K, Verron, M, Canard, B. | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Enzymatic Synthesis of Acyclic Nucleoside Thiophosphonate Diphosphates: Effect of the Alpha-Phosphorus Configuration on HIV-1 RT Activity.
Antiviral Res., 117, 2015
|
|
