5SQ9
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894420 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQ7
 
 | |
3FPD
 
 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | Authors: | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5SQH
 
 | |
5SQG
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894430 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5016127255 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[4-(methylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SRV
 
 | |
3IQA
 
 | | Crystal Structure of BlaC covalently bound with Doripenem | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2009-08-19 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem.
Biochemistry, 49, 2010
|
|
5SQF
 
 | |
5SQK
 
 | |
6KPC
 
 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
5SQJ
 
 | |
5X38
 
 | |
5SQN
 
 | |
5X36
 
 | |
5X3C
 
 | |
4XYO
 
 | | Structure of AgrA LytTR domain | | Descriptor: | 1,2-ETHANEDIOL, Accessory gene regulator A | | Authors: | Gopal, B, Rajasree, K. | | Deposit date: | 2015-02-02 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational features of theStaphylococcus aureusAgrA-promoter interactions rationalize quorum-sensing triggered gene expression.
Biochem Biophys Rep, 6, 2016
|
|
5X37
 
 | |
5SPO
 
 | |
4BFX
 
 | | Crystal structure of Mycobacterium tuberculosis PanK in complex with a triazole inhibitory compound (1f) and phosphate | | Descriptor: | 2,6-difluoro-N-[1-(5-{[2-(4-fluorophenoxy)ethyl]sulfanyl}-4-methyl-4H-1,2,4-triazol-3-yl)ethyl]benzamide, PANTOTHENATE KINASE, PHOSPHATE ION | | Authors: | Bjorkelid, C, Bergfors, T, Jones, T.A. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Characterization of Compounds Inhibiting Mycobacterium Tuberculosis Pank
J.Biol.Chem., 288, 2013
|
|
5SPR
 
 | |
5SFV
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(c(c(C(N1CCC1)=O)cn2)C(Nc4nn(C3CCCC3)cc4)=O)C, micromolar IC50=0.075252 | | Descriptor: | 4-(azetidine-1-carbonyl)-N-(1-cyclopentyl-1H-pyrazol-3-yl)-1-methyl-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
3IUO
 
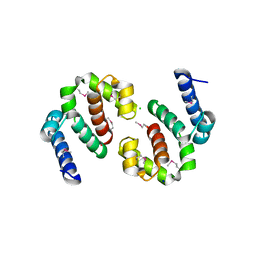 | | The Crystal Structure of the C-terminal domain of the ATP-dependent DNA helicase RecQ from Porphyromonas gingivalis to 1.6A | | Descriptor: | ATP-dependent DNA helicase RecQ, CHLORIDE ION, SODIUM ION | | Authors: | Stein, A.J, Sather, A, Duggan, E, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the C-terminal domain of the ATP-dependent DNA helicase RecQ from Porphyromonas gingivalis to 1.6A
To be Published
|
|
5SFZ
 
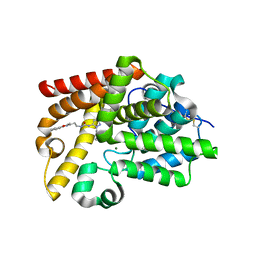 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(ccnn1c2ccccc2)NC(=O)NCCc3c(oc(n3)c4ccccc4)C, micromolar IC50=0.843056 | | Descriptor: | MAGNESIUM ION, N-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethyl]-N'-(1-phenyl-1H-pyrazol-5-yl)urea, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SPU
 
 | |
