2YF4
 
 | | Crystal structure of DR2231, the MazG-like protein from Deinococcus radiodurans, Apo structure | | Descriptor: | GLYCEROL, MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE, SULFATE ION | | Authors: | Goncalves, A.M.D, deSanctis, D, McSweeney, S.M. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
2WB4
 
 | | activated diguanylate cyclase PleD in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), BERYLLIUM TRIFLUORIDE ION, DIGUANYLATE CYCLASE, ... | | Authors: | Wassmann, P, Schirmer, T. | | Deposit date: | 2009-02-20 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Activated Pled, Identification of Dimerization and Catalysis Relevant Regulatory Mechanisms
To be Published
|
|
4RGW
 
 | | Crystal Structure of a TAF1-TAF7 Complex in Human Transcription Factor IID | | Descriptor: | GLYCEROL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7 | | Authors: | Wang, H, Curran, E.C, Hinds, T.R, Wang, E.H, Zheng, N. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module.
Cell Res., 24, 2014
|
|
3G2Z
 
 | |
8DLW
 
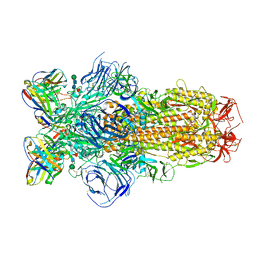 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with Fab S2M11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab S2M11 heavy chain, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
3ZK7
 
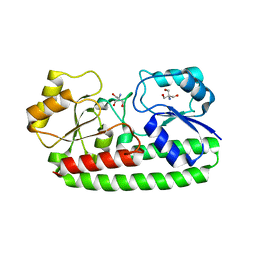 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA IN THE METAL-FREE, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
6IJA
 
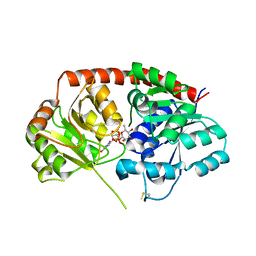 | | Crystal Structure of Arabidopsis thaliana UGT89C1 complexed with UDP-L-rhamnose | | Descriptor: | UDP-glycosyltransferase 89C1, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Zong, G, Wang, X. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-beta-l-rhamnose and rhamnosylation mechanism.
Plant J., 99, 2019
|
|
7HF3
 
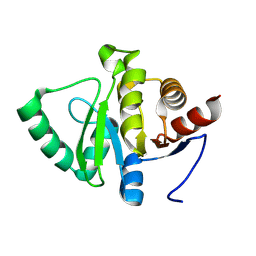 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0004064 | | Descriptor: | Non-structural protein 3, [(2R,4S)-4-methyl-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-2-yl]methanol, [(2S,4R)-4-methyl-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-2-yl]methanol | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
7MM6
 
 | |
7HCB
 
 | |
7HCK
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0000329 | | Descriptor: | (2S)-6-methyl-N-[(4S)-5,6,7,8-tetrahydro[1,2,4]triazolo[4,3-a]pyridin-3-yl]-3,4-dihydro-2H-1-benzopyran-2-carboxamide, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
3ZU3
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (MR, cleaved Histag) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
7MMH
 
 | |
7HCC
 
 | |
7HCJ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0000328 | | Descriptor: | (2R)-2-methyl-3-[(2-oxo-1,2,3,4-tetrahydroquinoline-6-carbonyl)amino]propanoic acid, (2S)-2-methyl-3-[(2-oxo-1,2,3,4-tetrahydroquinoline-6-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
6ABZ
 
 | | Crystal Structure of HEWL in deionized water | | Descriptor: | CHLORIDE ION, Lysozyme C, S-1,2-PROPANEDIOL, ... | | Authors: | Seyedarabi, A, Seraj, Z. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The role of Cinnamaldehyde and Phenyl ethyl alcohol as two types of precipitants affecting protein hydration levels.
Int.J.Biol.Macromol., 146, 2019
|
|
4RIC
 
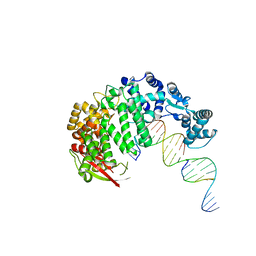 | | FAN1 Nuclease bound to 5' hydroxyl (dT-dT) single flap DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*AP*GP*GP*AP*GP*TP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*CP*CP*AP*CP*GP*CP*CP*TP*AP*GP*AP*CP*TP*CP*CP*TP*C)-3'), ... | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
5CVA
 
 | | Crystal structure of the type IX collagen NC2 hetero-trimerization domain with a guest fragment a1a2a1 of type I collagen | | Descriptor: | Collagen alpha-1(I) chain,Collagen alpha-2(IX) chain, Collagen alpha-1(I) chain,Collagen alpha-3(IX) chain, Collagen alpha-2(I) chain,Collagen alpha-1(IX) chain, ... | | Authors: | Boudko, S.P, Bachinger, H.P. | | Deposit date: | 2015-07-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Structural insight for chain selection and stagger control in collagen.
Sci Rep, 6, 2016
|
|
4RI9
 
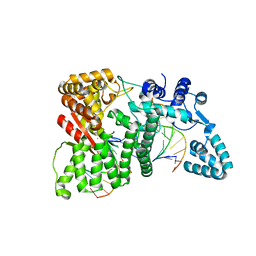 | |
3UEQ
 
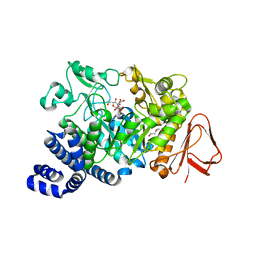 | | Crystal structure of amylosucrase from Neisseria polysaccharea in complex with turanose | | Descriptor: | 3-O-alpha-D-glucopyranosyl-D-fructose, Amylosucrase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Guerin, F, Pizzut-Serin, S, Potocki-Veronese, G, Guillet, V, Mourey, L, Remaud-Simeon, M, Andre, I, Tranier, S. | | Deposit date: | 2011-10-31 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Investigation of the Thermostability and Product Specificity of Amylosucrase from the Bacterium Deinococcus geothermalis.
J.Biol.Chem., 287, 2012
|
|
4O4D
 
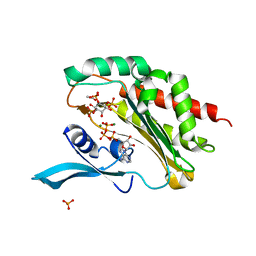 | | Crystal Structure of an Inositol hexakisphosphate kinase EhIP6KA in complexed with ATP and Ins(1,4,5)P3 | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family.
Nat Commun, 5, 2014
|
|
3MKB
 
 | | Crystal structure determination of Shortfin Mako (Isurus oxyrinchus) hemoglobin at 1.9 Angstrom resolution | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ramesh, P, Sundaresan, S.S, Sathya Moorthy, P, Balasubramanian, M, Ponnuswamy, M.N. | | Deposit date: | 2010-04-14 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of haemoglobin from pisces species shortfin mako shark (Isurus oxyrinchus) at 1.9 angstrom resolution
J.SYNCHROTRON RADIAT., 20, 2013
|
|
5CXX
 
 | | Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Gruninger, R.J, Abbott, D.W. | | Deposit date: | 2015-07-29 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Contributions of a unique beta-clamp to substrate recognition illuminates the molecular basis of exolysis in ferulic acid esterases.
Biochem.J., 473, 2016
|
|
5TWS
 
 | | Post-catalytic complex of human Polymerase Mu (H329A) with newly incorporated UTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
7HQW
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z431953146 | | Descriptor: | (5-phenyl-1,2-oxazol-3-yl)methanol, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
