4N5L
 
 | |
6O09
 
 | |
5X51
 
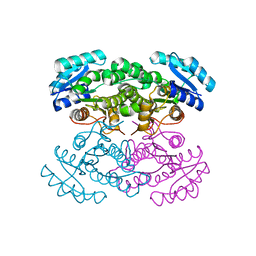
 | | RNA Polymerase II from Komagataella Pastoris (Type-3 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.996 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
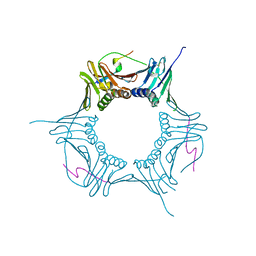
5X50
 
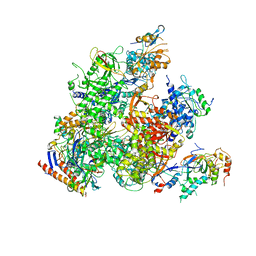 | | RNA Polymerase II from Komagataella Pastoris (Type-2 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.293 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
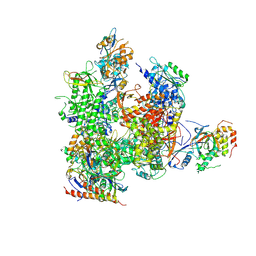
5X4Z
 
 | | RNA Polymerase II from Komagataella Pastoris (Type-1 crystal) | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, RNA polymerase II subunit, ... | | Authors: | Ehara, H, Umehara, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.8 Å) | | Cite: | Crystal structure of RNA polymerase II from Komagataella pastoris
Biochem. Biophys. Res. Commun., 487, 2017
|
|
3JUX
 
 | |
3JV2
 
 | | Crystal Structure of B. subtilis SecA with bound peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein translocase subunit SecA, ... | | Authors: | Zimmer, J. | | Deposit date: | 2009-09-15 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational flexibility and peptide interaction of the translocation ATPase SecA.
J.Mol.Biol., 394, 2009
|
|
7BM8
 
 | | Crystal structure of the C-terminally truncated chromosome-partitioning protein ParB from Caulobacter crescentus complexed with CTP-gamma-S | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Chromosome-partitioning protein ParB, MAGNESIUM ION | | Authors: | Jalal, A.S, Tran, N.T, Stevenson, C.E.M, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2021-01-19 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A CTP-dependent gating mechanism enables ParB spreading on DNA.
Elife, 10, 2021
|
|
5TBZ
 
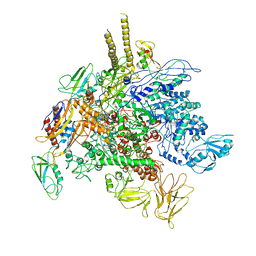 | | E. Coli RNA Polymerase complexed with NusG | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Steitz, T.A. | | Deposit date: | 2016-09-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Structural insights into NusG regulating transcription elongation.
Nucleic Acids Res., 45, 2017
|
|
3CGU
 
 | |
1I3Q
 
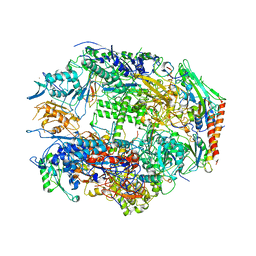 | | RNA POLYMERASE II CRYSTAL FORM I AT 3.1 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-15 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
1I50
 
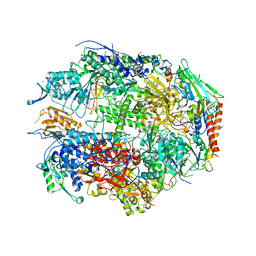 | | RNA POLYMERASE II CRYSTAL FORM II AT 2.8 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-23 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
1NT9
 
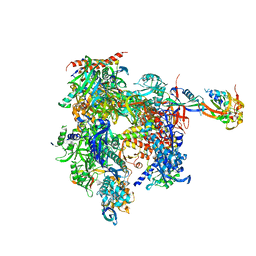 | | Complete 12-subunit RNA polymerase II | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6 KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 19 KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II LARGEST SUBUNIT, ... | | Authors: | Armache, K.-J, Kettenberger, H, Cramer, P. | | Deposit date: | 2003-01-29 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Architecture of initiation-competent 12-subunit RNA polymerase II
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1Q9I
 
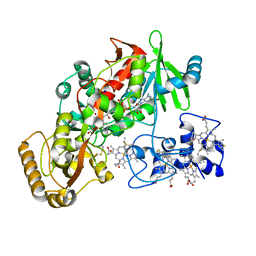 | | The A251C:S430C double mutant of flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEME C, MALATE LIKE INTERMEDIATE, ... | | Authors: | Rothery, E.L, Mowat, C.G, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Domain Mobility in a Flavocytochrome
Biochemistry, 42, 2004
|
|
7VUF
 
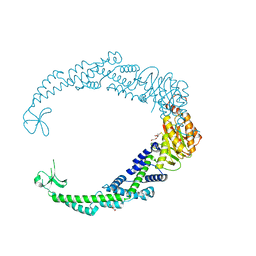 | |
7VUK
 
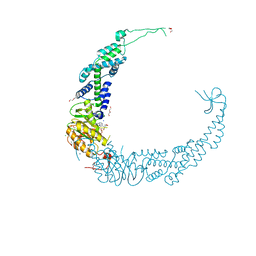 | |
4ATZ
 
 | |
6KLW
 
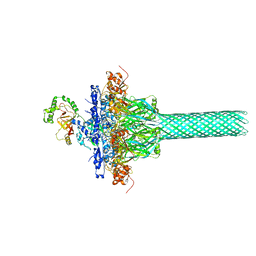 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1TZN
 
 | | Crystal Structure of the Anthrax Toxin Protective Antigen Heptameric Prepore bound to the VWA domain of CMG2, an anthrax toxin receptor | | Descriptor: | Anthrax toxin receptor 2, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Lacy, D.B, Wigelsworth, D.J, Melnyk, R.A, Collier, R.J. | | Deposit date: | 2004-07-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TZO
 
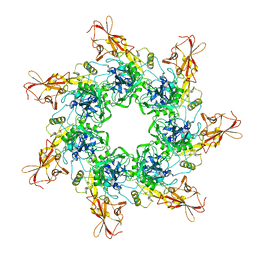 | | Crystal Structure of the Anthrax Toxin Protective Antigen Heptameric Prepore | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lacy, D.B, Wigelsworth, D.J, Melnyk, R.A, Collier, R.J. | | Deposit date: | 2004-07-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T6B
 
 | | Crystal structure of B. anthracis Protective Antigen complexed with human Anthrax toxin receptor | | Descriptor: | Anthrax toxin receptor 2, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Santelli, E, Bankston, L.A, Leppla, S.H, Liddington, R.C. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a complex between anthrax toxin and its host cell receptor
Nature, 430, 2004
|
|
1IZ5
 
 | | Pyrococcus furiosus PCNA mutant (Met73Leu, Asp143Ala, Asp147Ala): orthorhombic form | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-09-23 | | Release date: | 2003-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intermolecular ion pairs maintain the toroidal structure of Pyrococcus furiosus PCNA
PROTEIN SCI., 12, 2003
|
|
4HBN
 
 | | Crystal structure of the human HCN4 channel C-terminus carrying the S672R mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Xu, X, Marni, F, Wu, X, Su, Z, Musayev, F, Shrestha, S, Xie, C, Gao, W, Liu, Q, Zhou, L. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Local and Global Interpretations of a Disease-Causing Mutation near the Ligand Entry Path in Hyperpolarization-Activated cAMP-Gated Channel.
Structure, 20, 2012
|
|
4H2A
 
 | |
4JWX
 
 | | GluN2A ligand-binding core in complex with propyl-NHP5G | | Descriptor: | (2R)-amino(1-hydroxy-4-propyl-1H-pyrazol-5-yl)ethanoic acid, GluN2A | | Authors: | Hansen, K.B, Tajima, N, Risgaard, R, Perszyk, R.E, Jorgensen, L, Vance, K.M, Ogden, K.K, Clausen, R.P, Furukawa, H, Traynelis, S.F. | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-d-aspartate receptors.
Mol.Pharmacol., 84, 2013
|
|
