3PCM
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 6-HYDROXYNICOTINIC ACID N-OXIDE AND CYANIDE | | Descriptor: | 6-HYDROXYISONICOTINIC ACID N-OXIDE, CYANIDE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCA
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCI
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-IODO-4-HYDROXYBENZOATE | | Descriptor: | 3-IODO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCK
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 6-HYDROXYNICOTINIC ACID N-OXIDE | | Descriptor: | 6-HYDROXYISONICOTINIC ACID N-OXIDE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCJ
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 2-HYDROXYISONICOTINIC ACID N-OXIDE | | Descriptor: | 2-HYDROXYISONICOTINIC ACID N-OXIDE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCL
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 2-HYDROXYISONICOTINIC ACID N-OXIDE AND CYANIDE | | Descriptor: | 2-HYDROXYISONICOTINIC ACID N-OXIDE, CYANIDE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
1AOK
 
 | | VIPOXIN COMPLEX | | Descriptor: | ACETATE ION, VIPOXIN COMPLEX | | Authors: | Perbandt, M, Wilson, J.C, Eschenburg, S, Betzel, C. | | Deposit date: | 1997-07-07 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of vipoxin at 2.0 A: an example of regulation of a toxic function generated by molecular evolution.
FEBS Lett., 412, 1997
|
|
1AYN
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Oliveira, M.A, Zhao, R, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human rhinovirus 16.
Structure, 1, 1993
|
|
1MRP
 
 | | FERRIC-BINDING PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | FE (III) ION, FERRIC IRON BINDING PROTEIN, PHOSPHATE ION | | Authors: | Bruns, C.M, Nowalk, A.J, Arvai, A.S, Mctigue, M.A, Vaughan, K.G, Mietzner, T.A, Mcree, D.E. | | Deposit date: | 1997-05-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Haemophilus influenzae Fe(+3)-binding protein reveals convergent evolution within a superfamily.
Nat.Struct.Biol., 4, 1997
|
|
1DFG
 
 | | X-RAY STRUCTURE OF ESCHERICHIA COLI ENOYL REDUCTASE WITH BOUND NAD AND BENZO-DIAZABORINE | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ENOYL ACYL CARRIER PROTEIN REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baldock, C, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1997-01-16 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mechanism of drug action revealed by structural studies of enoyl reductase.
Science, 274, 1996
|
|
1NFD
 
 | | AN ALPHA-BETA T CELL RECEPTOR (TCR) HETERODIMER IN COMPLEX WITH AN ANTI-TCR FAB FRAGMENT DERIVED FROM A MITOGENIC ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H57 FAB, N15 ALPHA-BETA T-CELL RECEPTOR | | Authors: | Wang, J.-H, Lim, K, Smolyar, A, Teng, M.-K, Sacchittini, J, Reinherz, E.L. | | Deposit date: | 1997-08-04 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody.
EMBO J., 17, 1998
|
|
1TIP
 
 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, PHOSPHOENZYME INTERMEDIATE OF FRU-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.-H, Olson, T.W, Ogata, C.M, Levitt, D.G, Banaszak, L.J, Lange, A.J. | | Deposit date: | 1997-05-28 | | Release date: | 1998-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a trapped phosphoenzyme during a catalytic reaction.
Nat.Struct.Biol., 4, 1997
|
|
1AP8
 
 | | TRANSLATION INITIATION FACTOR EIF4E IN COMPLEX WITH M7GDP, NMR, 20 STRUCTURES | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, TRANSLATION INITIATION FACTOR EIF4E | | Authors: | Matsuo, H, Li, H, Mcguire, A.M, Fletcher, M, Gingras, A.C, Sonenberg, N, Wagner, G. | | Deposit date: | 1997-07-25 | | Release date: | 1998-01-28 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein.
Nat.Struct.Biol., 4, 1997
|
|
1AYI
 
 | | COLICIN E7 IMMUNITY PROTEIN IM7 | | Descriptor: | COLICIN E IMMUNITY PROTEIN 7 | | Authors: | Dennis, C.A, Pauptit, R.A, Wallis, R, James, R, Moore, G.R, Kleanthous, C. | | Deposit date: | 1997-11-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural comparison of the colicin immunity proteins Im7 and Im9 gives new insights into the molecular determinants of immunity-protein specificity.
Biochem.J., 333 ( Pt 1), 1998
|
|
1AP6
 
 | |
1IRF
 
 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERFERON REGULATORY FACTOR-2 | | Authors: | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | Deposit date: | 1997-11-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
4GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 Y108F MUTANT | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Oakley, A, Rossjohn, J, Parker, M. | | Deposit date: | 1997-01-20 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multifunctional role of Tyr 108 in the catalytic mechanism of human glutathione transferase P1-1. Crystallographic and kinetic studies on the Y108F mutant enzyme.
Biochemistry, 36, 1997
|
|
1ASK
 
 | | NUCLEAR TRANSPORT FACTOR 2 (NTF2) H66A MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Mccoy, A.J, Stewart, M.J. | | Deposit date: | 1997-08-11 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran.
J.Mol.Biol., 272, 1997
|
|
1JMG
 
 | |
1AXR
 
 | | COOPERATIVITY BETWEEN HYDROGEN-BONDING AND CHARGE-DIPOLE INTERACTIONS IN THE INHIBITION OF BETA-GLYCOSIDASES BY AZOLOPYRIDINES: EVIDENCE FROM A STUDY WITH GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 4,5,6-TRIHYDROXY-7-HYDROXYMETHYL-4,5,6,7-TETRAHYDRO-1H-[1,2,3]TRIAZOLO[1,5-A]PYRIDIN-8-YLIUM, GLYCOGEN PHOSPHORYLASE, PHOSPHATE ION, ... | | Authors: | Oikonomakos, N.G, Heightman, T.D. | | Deposit date: | 1997-10-20 | | Release date: | 1998-01-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cooperative interactions of the catalytic nucleophile and the catalytic acid in the inhibition of beta-glycosidases. Calculations and their validation by comparative kinetic and structural studies of the inhibition of glycogen phosphorylase b.
HELV.CHIM.ACTA, 81, 1998
|
|
1TVU
 
 | |
1FMT
 
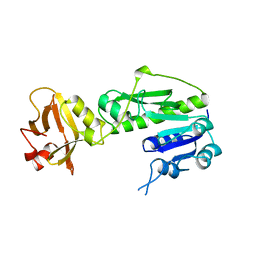 | | METHIONYL-TRNAFMET FORMYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA FMET FORMYLTRANSFERASE | | Authors: | Schmitt, E, Mechulam, Y. | | Deposit date: | 1997-10-13 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of crystalline Escherichia coli methionyl-tRNA(f)Met formyltransferase: comparison with glycinamide ribonucleotide formyltransferase.
EMBO J., 15, 1996
|
|
3CYR
 
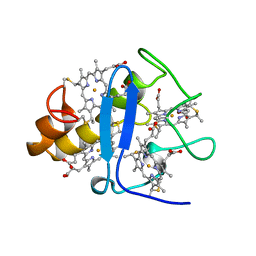 | | CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774P | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Simoes, P, Matias, P.M, Morais, J, Wilson, K, Dauter, Z, Carrondo, M.A. | | Deposit date: | 1997-07-24 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement of the Three-Dimensional Structures of Cytochrome C3 from Desulfovibrio Vulgaris Hildenborough at 1.67 Angstroms Resolution and from Desulfovibrio Desulfuricans Atcc 27774 at 1.6 Angstroms Resolution
Inorg.Chim.Acta., 273, 1998
|
|
1AX4
 
 | | TRYPTOPHANASE FROM PROTEUS VULGARIS | | Descriptor: | POTASSIUM ION, TRYPTOPHANASE | | Authors: | Isupov, M.N, Antson, A.A, Dodson, E.J, Dodson, G.G, Dementieva, I.S, Zakomirdina, L.N, Wilson, K.S, Dauter, Z, Lebedev, A.A, Harutyunyan, E.H. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of tryptophanase.
J.Mol.Biol., 276, 1998
|
|
1NWO
 
 | |
