7HLZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z256709358 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, SULFATE ION, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5ER0
 
 | |
5ERY
 
 | | Crystal Structure of APO MenD from M. tuberculosis - P212121 | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Johnston, J.M, Jirgis, E.N.M, Bashiri, G, Bulloch, E.M.M, Baker, E.N. | | Deposit date: | 2015-11-16 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Views along the Mycobacterium tuberculosis MenD Reaction Pathway Illuminate Key Aspects of Thiamin Diphosphate-Dependent Enzyme Mechanisms.
Structure, 24, 2016
|
|
7HNO
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z57450788 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-{4-[(morpholin-4-yl)methyl]phenyl}acetamide, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
9B9W
 
 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-40792 | | Descriptor: | (4P)-N-[(1R)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-[(25E)-1-{(1P)-6-fluoro-3'-[4-fluoro-2-(trifluoromethyl)benzamido]-4'-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-1,24-dioxo-5,8,11,14,17,20-hexaoxa-2,23-diazahexacos-25-en-26-yl]-1H-pyrrole-3-carboxamide, DDB1- and CUL4-associated factor 1, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Mamai, A, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
8BCA
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and compound 26 | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-~{N}-methyl-4-(methylamino)benzenesulfonamide, GLYCEROL, ... | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7HMK
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z271004858 | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-N-(pyridin-2-yl)benzenesulfonamide, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6MLS
 
 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from L-tyrosine | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-09-27 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
7HMU
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z1741966151 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-[(piperidin-4-yl)methyl]methanesulfonamide, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
8J00
 
 | | Human KCNQ2-CaM in complex with CBD | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, cannabidiol | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
9GX8
 
 | | Crystal structure of CIM-2, a membrane-bound B1 metallo-beta-lactamase from Chryseobacterium indologenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2024-09-28 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Active site loops of membrane-anchored metallo-beta-lactamases from environmental bacteria determine cephalosporinase activity.
Antimicrob.Agents Chemother., 69, 2025
|
|
9HGD
 
 | | Crystal structure of human GABARAP in complex with cyclic peptide GAB_D23 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Ueffing, A, Wilms, J.A, Willbold, D, Weiergraeber, O.H. | | Deposit date: | 2024-11-19 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Accurate de novo design of high-affinity protein-binding macrocycles using deep learning.
Nat.Chem.Biol., 2025
|
|
7VRJ
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF Allochromatium tepidum | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Tani, K, Kobayashi, K, Hosogi, N, Ji, X.-C, Nagashima, S, Nagashima, K.V.P, Tsukatani, Y, Kanno, R, Hall, M, Yu, L.-J, Ishikawa, I, Okura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-10-23 | | Release date: | 2022-05-04 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | A Ca 2+ -binding motif underlies the unusual properties of certain photosynthetic bacterial core light-harvesting complexes.
J.Biol.Chem., 298, 2022
|
|
7HNW
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z1446981563 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-[(9S)-6-oxaspiro[4.5]decan-9-yl]methanesulfonamide, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4O5T
 
 | | Crystal structure of Diels-Alderase CE20 in complex with a product analog | | Descriptor: | 4-{[2-(phosphonooxy)ethyl]carbamoyl}benzyl [(1R,6S)-6-(dimethylcarbamoyl)cyclohex-2-en-1-yl]carbamate, Diisopropyl-fluorophosphatase | | Authors: | Beck, T, Preiswerk, N, Mayer, C, Hilvert, D. | | Deposit date: | 2013-12-20 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Impact of scaffold rigidity on the design and evolution of an artificial Diels-Alderase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9B9H
 
 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-40333 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-{(1P)-5'-({(17E)-18-[(3P)-4-{[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]carbamoyl}-3-(4-chloro-2-fluorophenyl)-1H-pyrrol-2-yl]-16-oxo-3,6,9,12-tetraoxa-15-azaoctadec-17-en-1-yl}carbamoyl)-2'-fluoro-4-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Alvarez, H.G, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-02 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9HFY
 
 | | Crystal structure of SARS CoV-2 3CLpro (Mpro) with ALG-097078 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonyl]-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5 | | Authors: | Tauchert, M.J, Maskos, K, McGowan, D.C, Stoycheva, A.D. | | Deposit date: | 2024-11-18 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.282 Å) | | Cite: | Discovery and Preclinical Profile of ALG-097558, a Pan-Coronavirus 3CLpro Inhibitor.
J.Med.Chem., 68, 2025
|
|
8S38
 
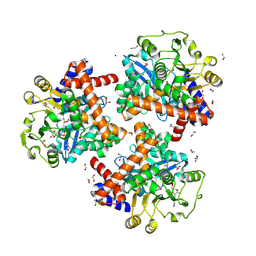 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 in complex with citrate and NAD | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
7HND
 
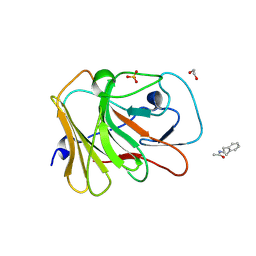 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z52314092 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, SULFATE ION, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5EGR
 
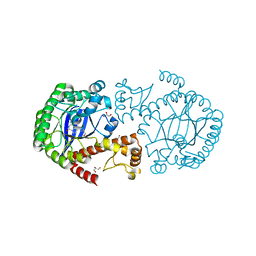 | | tRNA guanine transglycosylase (TGT) in complex with an Immucillin derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-7-[(2~{S},3~{R},5~{S})-5-(hydroxymethyl)-3-oxidanyl-pyrrolidin-2-yl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, GLYCEROL, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of an Immucillin Derivative as a Class of tRNA-Guanine Transglycosylase Inhibitors: Exploration of a Transition State Analogous Binding Mode
To be Published
|
|
8JH3
 
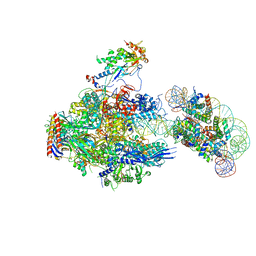 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8S3C
 
 | | Crystal structure of Medicago truncatula glutamate dehydrogenase 2 (unliganded) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2024-02-19 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Legume-type glutamate dehydrogenase: Structure, activity, and inhibition studies.
Int.J.Biol.Macromol., 278, 2024
|
|
5YUI
 
 | | CO2 release in human carbonic anhydrase II crystals: reveal histidine 64 and solvent dynamics | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Park, S.Y, McKenna, R. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7HMS
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z126932614 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(methylsulfonyl)methyl]-1H-benzimidazole, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6LUI
 
 | | Crystal structure of the SAMD1 WH domain and DNA complex | | Descriptor: | Atherin, DNA (5'-D(*AP*CP*CP*TP*GP*CP*GP*CP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*AP*TP*GP*GP*TP*GP*CP*GP*CP*AP*GP*GP*T)-3') | | Authors: | Zhou, Y, Cao, Y, Wang, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Sci Adv, 7, 2021
|
|
