8J0O
 
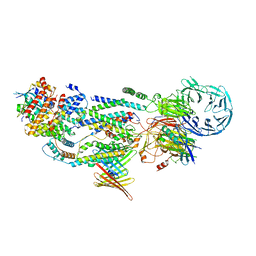 | | cryo-EM structure of human EMC and VDAC | | Descriptor: | ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ER membrane protein complex subunit 2, ... | | Authors: | Li, M, Zhang, C, Wu, J, Lei, M. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural insights into human EMC and its interaction with VDAC.
Aging (Albany NY), 16, 2024
|
|
5CAE
 
 | |
5DKO
 
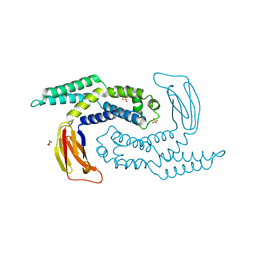 | | The structure of Escherichia coli ZapD | | Descriptor: | Cell division protein ZapD, SULFATE ION | | Authors: | Wroblewski, C, Kimber, M.S. | | Deposit date: | 2015-09-03 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mutational Analyses of Escherichia coli ZapD Reveal Charged Residues Involved in FtsZ Filament Bundling.
J.Bacteriol., 198, 2016
|
|
4QAC
 
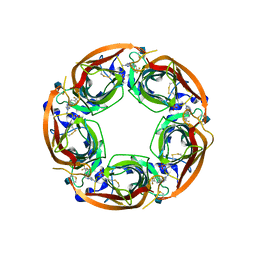 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 4-(4-methylpiperidin-1-yl)-6-(4-(trifluoromethyl)phenyl)pyrimidin-2-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(4-methylpiperidin-1-yl)-6-[4-(trifluoromethyl)phenyl]pyrimidin-2-amine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Radic, Z, Changeux, J.-P, Finn, M.G, Taylor, P. | | Deposit date: | 2014-05-03 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for cooperative interactions of substituted 2-aminopyrimidines with the acetylcholine binding protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7JFP
 
 | | GTP-specific succinyl-CoA synthetase complexed with Mg-GDP, phosphohistidine loop pointing towards nucleotide binding site | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Second distinct conformation of the phosphohistidine loop in succinyl-CoA synthetase
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JJ0
 
 | | GTP-specific succinyl-CoA synthetase complexed with Mg-GMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2020-07-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Second distinct conformation of the phosphohistidine loop in succinyl-CoA synthetase
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JMK
 
 | |
7JKR
 
 | | GTP-specific succinyl-CoA synthetase complexed with Mg-GMPPNP, phosphohistidine loop pointing towards nucleotide binding site | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2020-07-28 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Second distinct conformation of the phosphohistidine loop in succinyl-CoA synthetase
Acta Crystallogr.,Sect.D, 77, 2021
|
|
2FPP
 
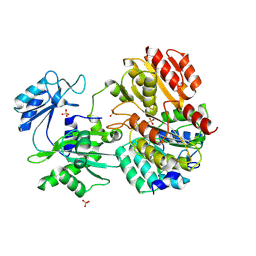 | | Crystal structure of pig GTP-specific succinyl-CoA synthetase from polyethylene glycol with chloride ions | | Descriptor: | CHLORIDE ION, SULFATE ION, Succinyl-CoA ligase [GDP-forming] alpha-chain, ... | | Authors: | Fraser, M.E, Hayakawa, K, Hume, M.S, Ryan, D.G, Brownie, E.R. | | Deposit date: | 2006-01-16 | | Release date: | 2006-02-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Interactions of GTP with the ATP-grasp Domain of GTP-specific Succinyl-CoA Synthetase
J.Biol.Chem., 281, 2006
|
|
7KUH
 
 | | MicroED structure of mVDAC | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Martynowycz, M.W, Khan, F, Hattne, J, Abramson, J, Gonen, T. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.12 Å) | | Cite: | MicroED structure of lipid-embedded mammalian mitochondrial voltage-dependent anion channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2FPG
 
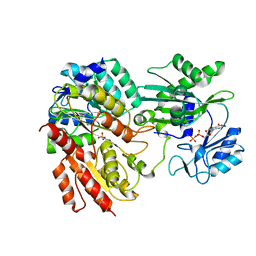 | |
2FP4
 
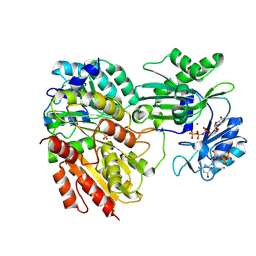 | |
2FPI
 
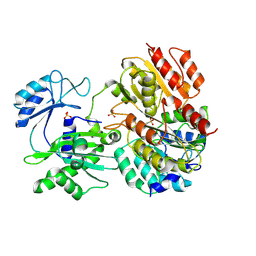 | |
7KZR
 
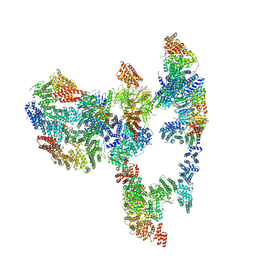 | | Structure of the human Fanconi Anaemia Core-UBE2T-ID complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KZP
 
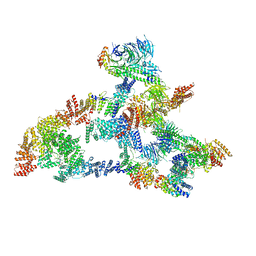 | | Structure of the human Fanconi anaemia Core complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
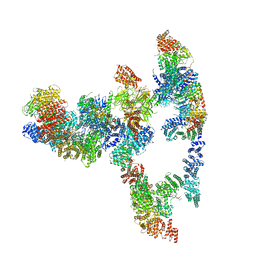
7KZV
 
 | |
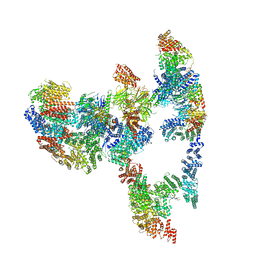
7KZT
 
 | |
7KZQ
 
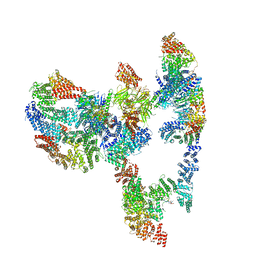 | | Structure of the human Fanconi anaemia Core-ID complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KZS
 
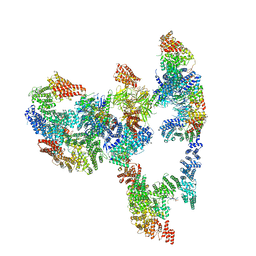 | |
2E2A
 
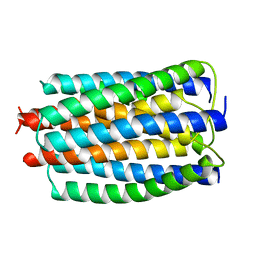 | |
2QLJ
 
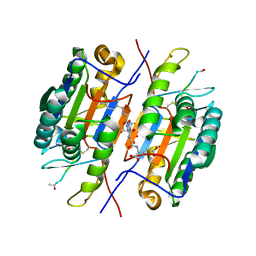 | | Crystal Structure of Caspase-7 with Inhibitor AC-WEHD-CHO | | Descriptor: | CITRIC ACID, Caspase-7, Inhibitor AC-WEHD-CHO, ... | | Authors: | Agniswamy, J, Fang, B, Weber, I. | | Deposit date: | 2007-07-12 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QL5
 
 | | Crystal Structure of caspase-7 with inhibitor AC-DMQD-CHO | | Descriptor: | CITRIC ACID, Caspase-7, inhibitor, ... | | Authors: | Agniswamy, J, Fang, B, Weber, I. | | Deposit date: | 2007-07-12 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QL9
 
 | | Crystal Structure of Caspase-7 with inhibitor AC-DQMD-CHO | | Descriptor: | CITRIC ACID, Caspase-7, Inhibitor AC-DQMD-CHO, ... | | Authors: | Agniswamy, J, Fang, B, Weber, I. | | Deposit date: | 2007-07-12 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
7WZS
 
 | |
3JZJ
 
 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose/maltose binding protein GacH, SULFATE ION | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-23 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
