4YFH
 
 | | HU38-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
7F0A
 
 | |
7F0B
 
 | | Crystal structure of capreomycin phosphotransferase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Capreomycin phosphotransferase | | Authors: | Chang, C.Y, Pan, Y.C, Wang, Y.L, Toh, S.I. | | Deposit date: | 2021-06-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dual-Mechanism Confers Self-Resistance to the Antituberculosis Antibiotic Capreomycin.
Acs Chem.Biol., 17, 2022
|
|
4YF0
 
 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
1FZZ
 
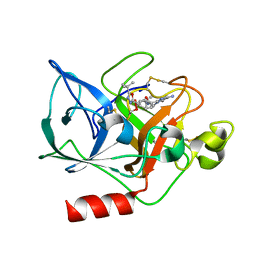 | | THE CRYSTAL STRUCTURE OF THE COMPLEX OF NON-PEPTIDIC INHIBITOR ONO-6818 AND PORCINE PANCREATIC ELASTASE. | | Descriptor: | 2-(5-AMINO-6-OXO-2-PHENYL-6H-PYRIMIDIN-1-YL)-N-[2-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)-1-(METHYLETHYL)-2-HYDROXYETHYL]ACETAMIDE, ELASTASE 1 | | Authors: | Odagaki, Y, Ohmoto, K, Matsuoka, S, Hamanaka, N, Nakai, H, Toda, M, Katsuya, Y. | | Deposit date: | 2000-10-04 | | Release date: | 2001-10-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The crystal structure of the complex of non-peptidic inhibitor of human neutrophil elastase ONO-6818 and porcine pancreatic elastase.
Bioorg.Med.Chem., 9, 2001
|
|
8FUM
 
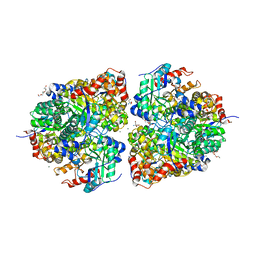 | | AibH1H2 metalated with Fe in the presence of Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
1PIS
 
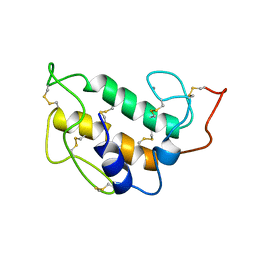 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B.D, Tessari, M, De Haas, G.H, Verheij, H.M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-12-22 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2.
EMBO J., 14, 1995
|
|
1PIR
 
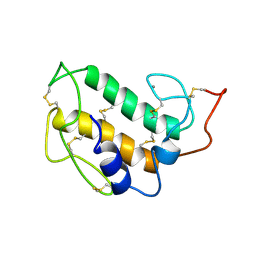 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PHOSPHOLIPASE A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B.D, Tessari, M, De Haas, G.H, Verheij, H.M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-12-22 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2.
EMBO J., 14, 1995
|
|
4YEX
 
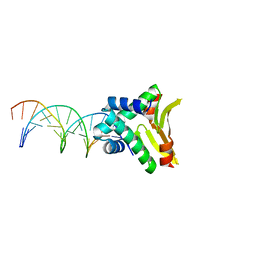 | | HUaa-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
5DOV
 
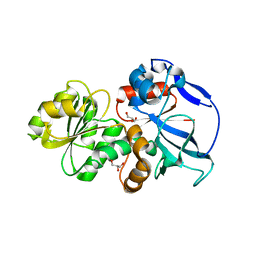 | |
1Q9E
 
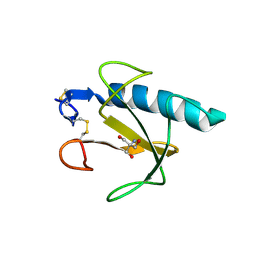 | | RNase T1 variant with adenine specificity | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Guanyl-specific ribonuclease T1 precursor | | Authors: | Czaja, R, Struhalla, M, Hoeschler, K, Saenger, W, Straeter, N, Hahn, U. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNase T1 Variant RV Cleaves Single-Stranded RNA after Purines Due to Specific Recognition by the Asn46 Side Chain Amide.
Biochemistry, 43, 2004
|
|
5DP1
 
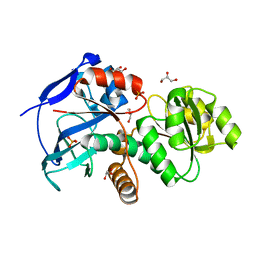 | | Crystal structure of CurK enoyl reductase | | Descriptor: | CurK, GLYCEROL, PHOSPHATE ION | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2015-09-12 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Cyclopropanation by a Unique Enoyl-Acyl Carrier Protein Reductase.
Structure, 23, 2015
|
|
2AZD
 
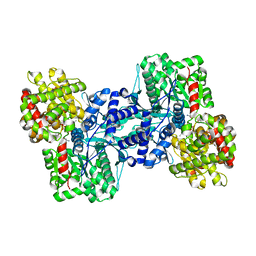 | |
1TZ4
 
 | | [hPP19-23]-pNPY bound to DPC Micelles | | Descriptor: | neuropeptide Y,Pancreatic prohormone,neuropeptide Y | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
5CBS
 
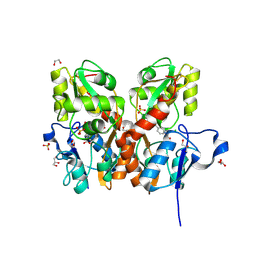 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid at 1.8A resolution | | Descriptor: | (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Studies on Aryl-Substituted Phenylalanines: Synthesis, Activity, and Different Binding Modes at AMPA Receptors.
J.Med.Chem., 59, 2016
|
|
6UNC
 
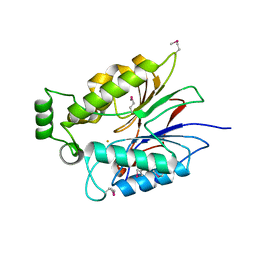 | |
7E35
 
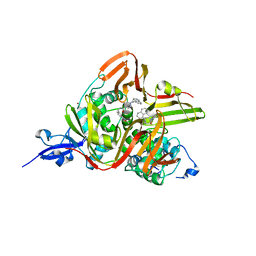 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
1HMC
 
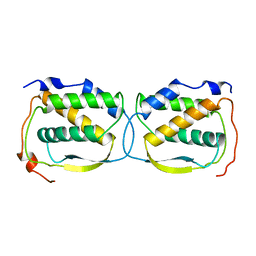 | | THREE-DIMENSIONAL STRUCTURE OF DIMERIC HUMAN RECOMBINANT MACROPHAGE COLONY STIMULATING FACTOR | | Descriptor: | HUMAN MACROPHAGE COLONY STIMULATING FACTOR | | Authors: | Bohm, A, Pandit, J, Jancarik, J, Halenbeck, R, Koths, K, Kim, S.-H. | | Deposit date: | 1993-12-22 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of dimeric human recombinant macrophage colony-stimulating factor.
Science, 258, 1992
|
|
8UIA
 
 | |
5OQ4
 
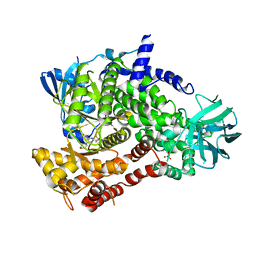 | | PQR309 - a Potent, Brain-Penetrant, Orally Bioavailable, pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology | | Descriptor: | 5-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Williams, R.L, Zhang, X. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology.
J. Med. Chem., 60, 2017
|
|
2DE9
 
 | |
5AKY
 
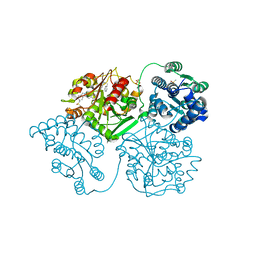 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 3-(2-phenylethyl)-1H-indazole, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5AK5
 
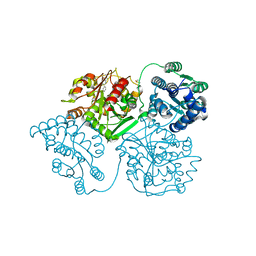 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 3-PROPYL-5-(3-PYRIDYL)-1H-PYRAZIN-2-ONE, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5AKH
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 4-[5-[3-(trifluoromethyl)phenyl]-1H-pyrazol-4-yl]pyridine, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
6UVM
 
 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W.C.K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
