3GWS
 
 | | Crystal Structure of T3-Bound Thyroid Hormone Receptor | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor beta | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Polikarpov, I, Baxter, J.D, Webb, P. | | Deposit date: | 2009-04-01 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function
J.Mol.Biol., 360, 2006
|
|
2Q6J
 
 | | Crystal Structure of Estrogen Receptor alpha Complexed to a B-N Substituted Ligand | | Descriptor: | 4-[(DIMESITYLBORYL)(2,2,2-TRIFLUOROETHYL)AMINO]PHENOL, Estrogen receptor, GRIP peptide | | Authors: | Zhou, H, Nettles, K.W, Bruning, J.B, Kim, Y, Joachimiak, A, Sharma, S, Carlson, K.E, Stossi, F, Katzenellenbogen, B.S, Greene, G.L, Katzenellenbogen, J.A. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Elemental isomerism: a boron-nitrogen surrogate for a carbon-carbon double bond increases the chemical diversity of estrogen receptor ligands
Chem.Biol., 14, 2007
|
|
7CX7
 
 | | Crystal structure of Arabinose isomerase from hybrid AI8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, L-arabinose isomerase, ... | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) wt
To Be Published
|
|
2PYI
 
 | | Crystal structure of Glycogen Phosphorylase in complex with glucosyl triazoleacetamide | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Alexacou, K.M, Tiraidis, C, Zographos, S.E, Chrysina, E.D, Hayes, J, Oikonomakos, N.G. | | Deposit date: | 2007-05-16 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystallographic and computational studies on 4-phenyl-N-(beta-D-glucopyranosyl)-1H-1,2,3-triazole-1-acetamide, an inhibitor of glycogen phosphorylase: Comparison with alpha-D-glucose, N-acetyl-beta-D-glucopyranosylamine and N-benzoyl-N'-beta-D-glucopyranosyl urea binding.
Proteins, 71, 2007
|
|
7CKE
 
 | |
7CKD
 
 | |
5FMS
 
 | | mmIFT52 N-terminal domain | | Descriptor: | INTRAFLAGELLAR TRANSPORT PROTEIN 52 HOMOLOG | | Authors: | Mourao, A, Lorentzen, E. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.488 Å) | | Cite: | Intraflagellar Transport Proteins 172, 80, 57, 54, 38, and 20 Form a Stable Tubulin-Binding Ift-B2 Complex.
Embo J., 35, 2016
|
|
5HJE
 
 | |
7D0I
 
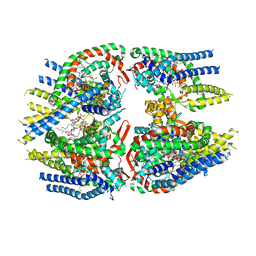 | | Cryo-EM structure of Schizosaccharomyces pombe Atg9 | | Descriptor: | Autophagy-related protein 9, Lauryl Maltose Neopentyl Glycol | | Authors: | Matoba, K, Tsutsumi, A, Kikkawa, M, Noda, N.N. | | Deposit date: | 2020-09-10 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2PIN
 
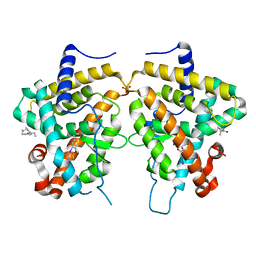 | | Thyroid receptor beta in complex with inhibitor | | Descriptor: | 1-(4-HEXYLPHENYL)PROP-2-EN-1-ONE, SULFATE ION, Thyroid hormone receptor beta-1, ... | | Authors: | Estebanez-Perpina, E, Jouravel, N, Baxter, J.D, Guy, L.R, Webb, P, Fletterick, R.J. | | Deposit date: | 2007-04-13 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the mode of action of a direct inhibitor of coregulator binding to the thyroid hormone receptor.
Mol.Endocrinol., 21, 2007
|
|
7DEN
 
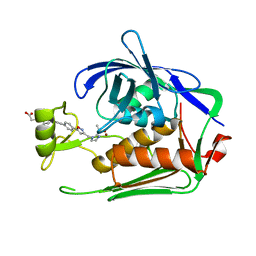 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 4-[(1~{R},5~{S})-6-[2-[4-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]phenyl]ethynyl]-3-azabicyclo[3.1.0]hexan-3-yl]butanoic acid, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Takashima, H. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
7DEL
 
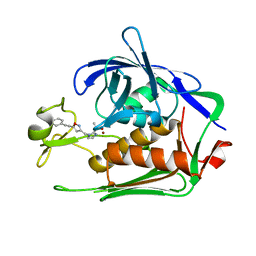 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 3-[4-[2-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]ethynyl]phenyl]propan-1-ol, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Tanaka-Yamamoto, N. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
7DEM
 
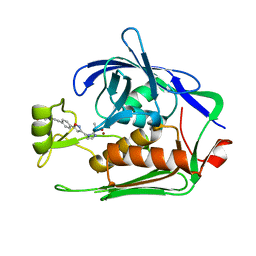 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 5-[4-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]phenyl]pent-4-yn-1-ol, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Matsuda, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
2PYS
 
 | | Crystal Structure of a Five Site Mutated Cyanovirin-N with a Mannose Dimer Bound at 1.8 A Resolution | | Descriptor: | Cyanovirin-N, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Fromme, R, Katilene, Z, Fromme, P, Ghirlanda, G. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Monovalent Mutant of Cyanovirin-N Provides Insight into the Role of Multiple Interactions with gp120 for Antiviral Activity.
Biochemistry, 46, 2007
|
|
3Q33
 
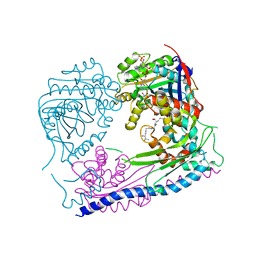 | | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, HISTONE H3, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
2Q7T
 
 | |
5XIQ
 
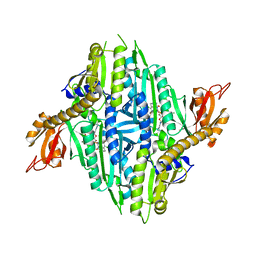 | | Crystal Structure of Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in complex with Halofuginone | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
5XM0
 
 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3.3, and H4 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.874 Å) | | Cite: | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
5HGC
 
 | | A Serpin structure | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, SULFATE ION, ... | | Authors: | Das, S, Vashchenko, G.V, Van Petegem, F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A Serpin structure
J.Biol.Chem., 2016
|
|
5XII
 
 | | Crystal Structure of Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in complex with inhibitor 6 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-7-fluoranyl-3-[3-[(2R)-3-oxidanylidenepiperidin-2-yl]propyl]quinazolin-4-one, CHLORIDE ION, ... | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
3NBF
 
 | | Q28E mutant of hera helicase N-terminal domain bound to 8-oxo-ADP | | Descriptor: | Heat resistant RNA dependent ATPase, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl dihydrogen phosphate, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl phosphono hydrogen phosphate | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
6A9E
 
 | | Crystal structure of the N-terminal domain of Atg2 | | Descriptor: | Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6AEF
 
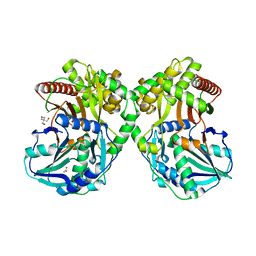 | | PapA2 acyl transferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Trehalose-2-sulfate acyltransferase PapA2, ... | | Authors: | Chaudhary, S, Rao, V, Panchal, V. | | Deposit date: | 2018-08-04 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A novel mutation alters the stability of PapA2 resulting in the complete abrogation of sulfolipids in clinical mycobacterial strains.
Faseb Bioadv, 1, 2019
|
|
6A9J
 
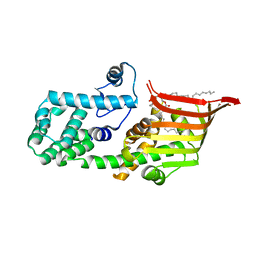 | | Crystal structure of the PE-bound N-terminal domain of Atg2 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
5ZSK
 
 | |
