1UYI
 
 | | Human Hsp90-alpha with 8-(2,5-dimethoxy-benzyl)-2-fluoro-9-pent-9H-purin-6-ylamine | | Descriptor: | 8-(2,5-DIMETHOXY-BENZYL)-2-FLUORO-9-PENT-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1UN2
 
 | | Crystal structure of circularly permuted CPDSBA_Q100T99: Preserved Global Fold and Local Structural Adjustments | | Descriptor: | THIOL-DISULFIDE INTERCHANGE PROTEIN | | Authors: | Manjasetty, B.A, Hennecke, J, Glockshuber, R, Heinemann, U. | | Deposit date: | 2003-09-03 | | Release date: | 2003-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Circularly Permuted Dsba(Q100T99): Preserved Global Fold and Local Structural Adjustments
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UPI
 
 | | Mycobacterium tuberculosis rmlC epimerase (Rv3465) | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE | | Authors: | Kim, C.-Y, Naranjo, C, Waldo, G.S, Lekin, T, Segelke, B.W, Kantardjieff, K.A, Zemla, A, Terwilliger, T, Rupp, B, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-10-05 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mycobacterium Tuberculosis Rmlc Epimerase (Rv3465): A Promising Drug-Target Structure in the Rhamnose Pathway
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UYF
 
 | | Human Hsp90-alpha with 8-(2-chloro-3,4,5-trimethoxy-benzyl)-2-fluoro-9-pent-4-ylnyl-9H-purin-6-ylamine | | Descriptor: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1UWU
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with D-glucohydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
1UYM
 
 | | Human Hsp90-beta with PU3 (9-Butyl-8(3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine) | | Descriptor: | 9-BUTYL-8-(3,4,5-TRIMETHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-BETA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1UYH
 
 | | Human Hsp90-alpha with 9-Butyl-8-(2,5-dimethoxy-benzyl)-2-fluoro-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(2,5-DIMETHOXY-BENZYL)-2-FLUORO-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1UWQ
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus | | Descriptor: | ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
2EYV
 
 | | SH2 domain of CT10-Regulated Kinase | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2EYW
 
 | |
1U6I
 
 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.2A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U9F
 
 | |
2FCV
 
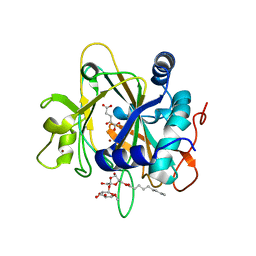 | | SyrB2 with Fe(II), bromide, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, BROMIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2FB3
 
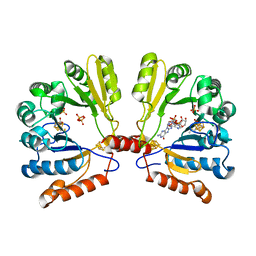 | | Structure of MoaA in complex with 5'-GTP | | Descriptor: | 5'-DEOXYADENOSINE, GUANOSINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2FFG
 
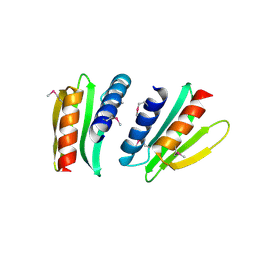 | | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360. | | Descriptor: | ykuJ | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Vorobiev, S.M, Ho, C.K, Janjua, H, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360.
To be Published
|
|
1U3T
 
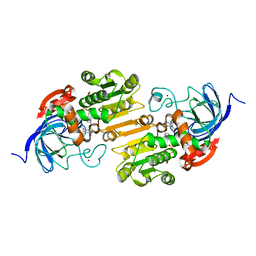 | |
2FCT
 
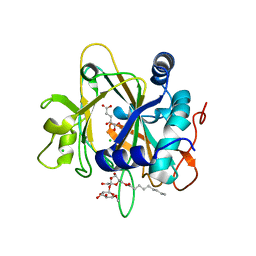 | | SyrB2 with Fe(II), chloride, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2FHD
 
 | | Crystal structure of Crb2 tandem tudor domains | | Descriptor: | DNA repair protein rhp9/CRB2, PHOSPHATE ION | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2005-12-23 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
1TVC
 
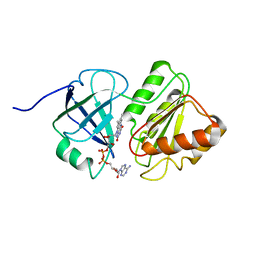 | | FAD and NADH binding domain of methane monooxygenase reductase from Methylococcus capsulatus (Bath) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, METHANE MONOOXYGENASE COMPONENT C | | Authors: | Chatwood, L.L, Mueller, J, Gross, J.D, Wagner, G, Lippard, S.J. | | Deposit date: | 2004-06-29 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Flavin Domain from Soluble Methane Monooxygenase Reductase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
2FLO
 
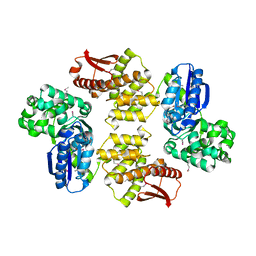 | |
1UC9
 
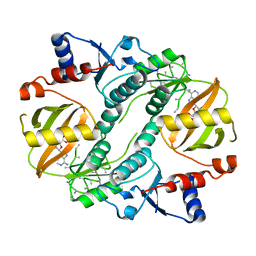 | | Crystal structure of a lysine biosynthesis enzyme, Lysx, from thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, lysine biosynthesis enzyme | | Authors: | Sakai, H, Vassylyeva, M.N, Matsuura, T, Sekine, S, Nishiyama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of a Lysine Biosynthesis Enzyme, LysX, from Thermus thermophilus HB8
J.Mol.Biol., 332, 2003
|
|
2DK4
 
 | | Solution structure of Splicing Factor Motif in Pre-mRNA splicing factor 18 (hPRP18) | | Descriptor: | Pre-mRNA-splicing factor 18 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the splicing factor motif of the human Prp18 protein.
Proteins, 80, 2012
|
|
1TQ7
 
 | | Crystal structure of the anticoagulant thrombin mutant W215A/E217A bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Prothrombin, ... | | Authors: | Pineda, A.O, Chen, Z.-W, Caccia, S, Savvides, S.N, Waksman, G, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket
J.Biol.Chem., 279, 2004
|
|
2DND
 
 | | A BIFURCATED HYDROGEN-BONDED CONFORMATION IN THE D(A.T) BASE PAIRS OF THE DNA DODECAMER D(CGCAAATTTGCG) AND ITS COMPLEX WITH DISTAMYCIN | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Coll, M, Frederick, C.A, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin.
Proc.Natl.Acad.Sci.USA, 84, 1987
|
|
1TSJ
 
 | |
