6UZD
 
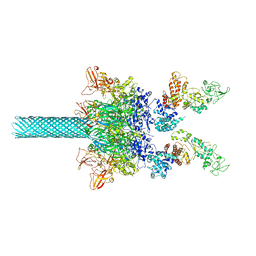 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
6UZE
 
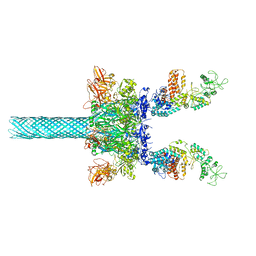 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
6DM6
 
 | |
6ZXK
 
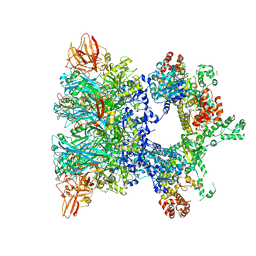 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state and PA7LF(2+1B) arrangement | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
6ZXL
 
 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state and PA7LF(2+1A) arrangement | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
6ZXJ
 
 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state, in which the third lethal factor is masked out (PA7LF3-masked) | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
3J9C
 
 | | CryoEM single particle reconstruction of anthrax toxin protective antigen pore at 2.9 Angstrom resolution | | Descriptor: | CALCIUM ION, Protective antigen PA-63 | | Authors: | Jiang, J, Pentelute, B.L, Collier, R.J, Zhou, Z.H. | | Deposit date: | 2014-12-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of anthrax protective antigen pore elucidates toxin translocation.
Nature, 521, 2015
|
|
6I5F
 
 | | Crystal structure of DNA-free E.coli MutS P839E dimer mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, GLYCEROL, ... | | Authors: | Bhairosing-Kok, D, Groothuizen, F.S, Fish, A, Dharadhar, S, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2018-11-13 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sharp kinking of a coiled-coil in MutS allows DNA binding and release.
Nucleic Acids Res., 47, 2019
|
|
3KWV
 
 | |
5OKI
 
 | |
5OKC
 
 | |
6T7X
 
 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6VRA
 
 | | Anthrax octamer prechannel bound to full-length edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
7YPE
 
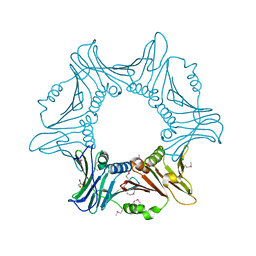 | |
7YPF
 
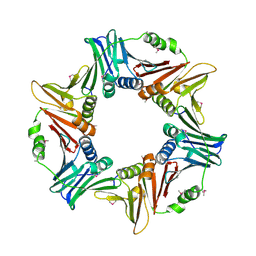 | |
1M2Z
 
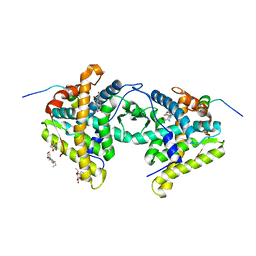 | | Crystal structure of a dimer complex of the human glucocorticoid receptor ligand-binding domain bound to dexamethasone and a TIF2 coactivator motif | | Descriptor: | DEXAMETHASONE, glucocorticoid receptor, nuclear receptor coactivator 2, ... | | Authors: | Bledsoe, R.B, Montana, V.G, Stanley, T.B, Delves, C.J, Apolito, C.J, Mckee, D.D, Consler, T.G, Parks, D.J, Stewart, E.L, Willson, T.M, Lambert, M.H, Moore, J.T, Pearce, K.H, Xu, H.E. | | Deposit date: | 2002-06-26 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Reveals a Novel Mode of Receptor Dimerization and Coactivator Recognition
Cell(Cambridge,Mass.), 110, 2002
|
|
7O1E
 
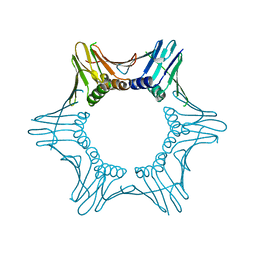 | | Crystal structure of PCNA from Chaetomium thermophilum | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Alphey, M.A, MacNeill, S, Yang, D. | | Deposit date: | 2021-03-29 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Non-canonical binding of the Chaetomium thermophilum PolD4 N-terminal PIP motif to PCNA involves Q-pocket and compact 2-fork plug interactions but no 3 10 helix.
Febs J., 290, 2023
|
|
7O1F
 
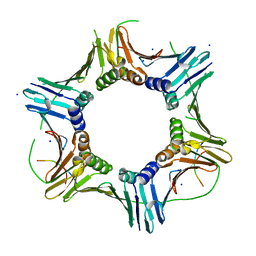 | |
1PLQ
 
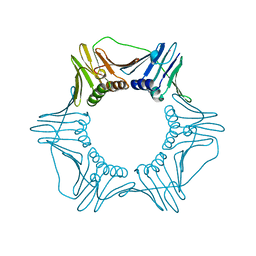 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | MERCURY (II) ION, PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
1PLR
 
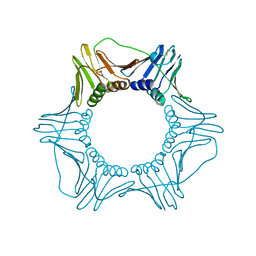 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
1S32
 
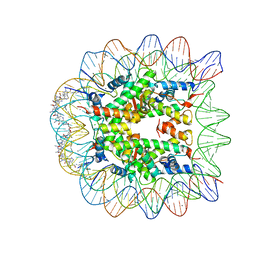 | | Molecular Recognition of the Nucleosomal 'Supergroove' | | Descriptor: | 2-(2-CARBAMOYLMETHOXY-ETHOXY)-ACETAMIDE, 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, ... | | Authors: | Edayathumangalam, R.S, Weyermann, P, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2004-01-12 | | Release date: | 2004-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Recognition of the Nucleosomal 'Supergroove'
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RXV
 
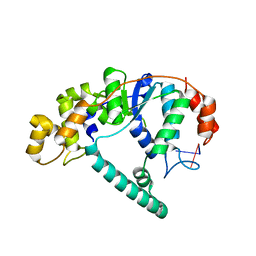 | | Crystal Structure of A. Fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG), Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXW
 
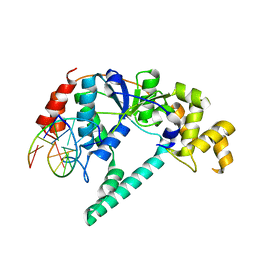 | | Crystal structure of A. fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*C*pG*pA*pT*pG*pC*pT*pA)-3', 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG)-3', Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
3A2F
 
 | |
5AUJ
 
 | |
