7QXU
 
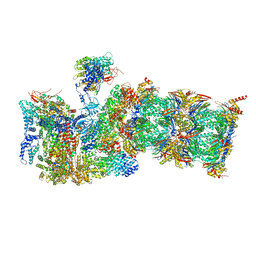 | | Proteasome-ZFAND5 Complex Z+C state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
8X4I
 
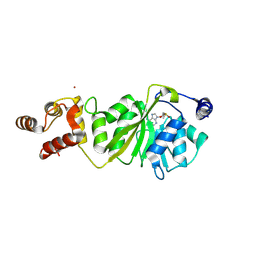 | |
7QXW
 
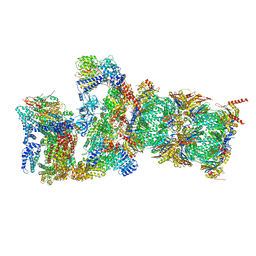 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
8X3W
 
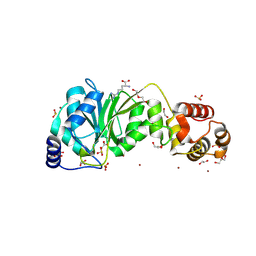 | | Crystal structure of DIMT1 from the thermophilic archaeon, Pyrococcus horikoshii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Sayan, S, Mandal, S.K, Dutta, A, Kanaujia, S.P. | | Deposit date: | 2023-11-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of archaeal DIMT1 unveils distinct protein dynamics essential for efficient catalysis.
Structure, 32, 2024
|
|
8X4G
 
 | |
8X44
 
 | |
7QYA
 
 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
8X4L
 
 | |
8X4P
 
 | |
8X4O
 
 | |
8X46
 
 | |
8X41
 
 | | Crystal structure of DIMT1 in complex with 5'-methylthioadenosine and adenosine from Pyrococcus horikoshii | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ACETATE ION, ... | | Authors: | Saha, S, Mandal, S.K, Dutta, A, Kanaujia, S.P. | | Deposit date: | 2023-11-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional characterization of archaeal DIMT1 unveils distinct protein dynamics essential for efficient catalysis.
Structure, 32, 2024
|
|
7QXN
 
 | | Proteasome-ZFAND5 Complex Z+A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYB
 
 | | Proteasome-ZFAND5 Complex Z-C state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
8WKW
 
 | | Structure of MAVS-CARD Filament | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Shi, M, Gao, P. | | Deposit date: | 2023-09-28 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Nucleic-acid-induced ZCCHC3 condensation promotes broad innate immune responses.
Mol.Cell, 85, 2025
|
|
8IUF
 
 | | Cryo-EM structure of Euglena gracilis super-complex I+III2+IV, composite | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Tian, H.T, He, Z.X, Hu, Y.Q, Zhou, L. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
8J9J
 
 | | Cryo-EM structure of Euglena gracilis complex I, NADH state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2023-05-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
8J9I
 
 | | Cryo-EM structure of Euglena gracilis complex I, turnover state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2023-05-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
8J9H
 
 | | Cryo-EM structure of Euglena gracilis respiratory complex I, deactive state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Wu, M.C, He, Z.X, Tian, H.T, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2023-05-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
7RZ8
 
 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RZ6
 
 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist glutamate | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RYY
 
 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist glutamate | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RZA
 
 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RYZ
 
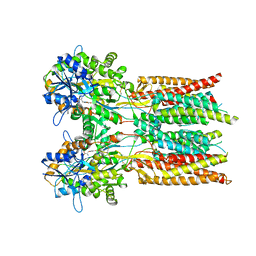 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RZ5
 
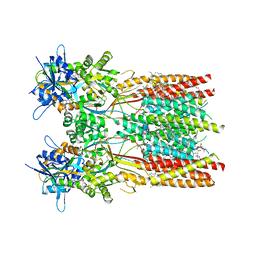 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK 200775 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
