1K7J
 
 | | Structural Genomics, protein TF1 | | Descriptor: | Protein yciO, SULFATE ION | | Authors: | Zhang, R, Dementieva, I, Thorn, J, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Genomics, protein TF1
To be Published
|
|
1ZS7
 
 | | The structure of gene product APE0525 from Aeropyrum pernix | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, hypothetical protein APE0525 | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-23 | | Release date: | 2005-07-05 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of gene product APE0525 from Aeropyrum pernix
To be Published
|
|
1ZSR
 
 | | Crystal structure of wild type HIV-1 protease (BRU isolate) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[S-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | N-{(2S,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-24 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | On the role of the R configuration of the reaction-intermediate isostere in HIV-1 protease-inhibitor binding: X-ray structure at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1XX6
 
 | | X-ray structure of Clostridium acetobutylicum thymidine kinase with ADP. Northeast Structural Genomics Target CAR26. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thymidine kinase, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Vorobiev, S.M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-11-04 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Clostridium acetobutylicum thymidine kinase with ADP. Northeast Structural Genomics Target CAR26.
To be Published
|
|
1XWY
 
 | | Crystal structure of tatD deoxyribonuclease from Escherichia coli K12 at 2.0 A resolution | | Descriptor: | Deoxyribonuclease tatD, ZINC ION | | Authors: | Malashkevich, V.N, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-11-02 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tatD DNase from Escherichia coli at 2.0 A resolution
To be Published
|
|
1XGF
 
 | |
1XPU
 
 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-dihydrobicyclomycin (FPDB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-(3-FORMYLPHENYLSULFANYL)-DIHYDROBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
1LRQ
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP, A5P and Cadmium | | Descriptor: | ARABINOSE-5-PHOSPHATE, CADMIUM ION, KDO-8-phosphate synthetase, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1XJ7
 
 | | Complex Androgen Receptor LBD and RAC3 peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, RAC3 derived peptide | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Buehrer, B.M, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-09-22 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
1JUB
 
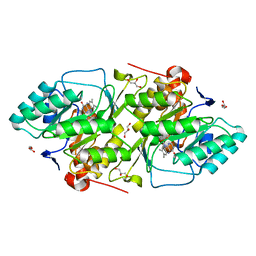 | | The K136E mutant of lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1XXL
 
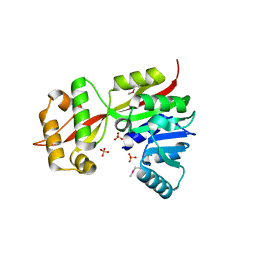 | |
1K04
 
 | |
1K2V
 
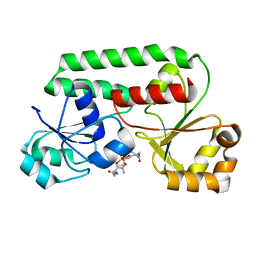 | | E. COLI PERIPLASMIC PROTEIN FHUD COMPLEXED WITH DESFERAL | | Descriptor: | DEFEROXAMINE MESYLATE FE(III) COMPLEX, Ferrichrome-binding periplasmic protein | | Authors: | Clarke, T.E, Braun, V, Winkelmann, G, Tari, L.W, Vogel, H.J. | | Deposit date: | 2001-09-29 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin.
J.Biol.Chem., 277, 2002
|
|
1JQX
 
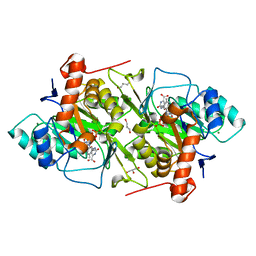 | | The R57A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JRC
 
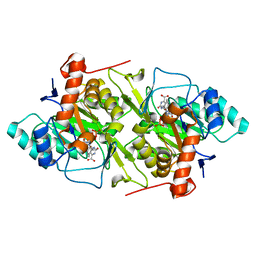 | | The N67A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, OROTIC ACID, dihydroorotate dehydrogenase A | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-13 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JOP
 
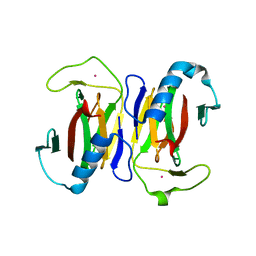 | |
1Y3J
 
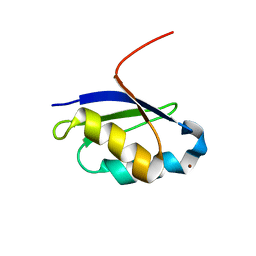 | | Solution structure of the copper(I) form of the fifth domain of Menkes protein | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Chasapis, C.T, Ciofi-Baffoni, S, Hadjiliadis, N, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-11-25 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal-binding domains of the Menkes protein
Febs J., 272, 2005
|
|
1ZZ0
 
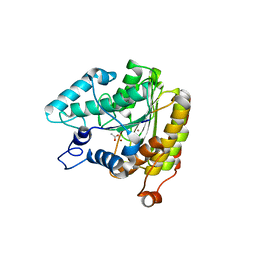 | | Crystal structure of a HDAC-like protein with acetate bound | | Descriptor: | ACETATE ION, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Dickmanns, A, Schwienhorst, A, Ficner, R. | | Deposit date: | 2005-06-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a bacterial class 2 histone deacetylase homologue
J.Mol.Biol., 354, 2005
|
|
220D
 
 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | BARIUM ION, DNA (5'-D(*AP*CP*CP*CP*GP*CP*GP*GP*GP*T)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
223D
 
 | | DIRECT OBSERVATION OF TWO BASE-PAIRING MODES OF A CYTOSINE-THYMINE ANALOGUE WITH GUANINE IN A DNA Z-FORM DUPLEX: SIGNIFICANCE FOR BASE ANALOGUE MUTAGENESIS | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C46)P*G)-3') | | Authors: | Moore, M.H, Van Meervelt, L, Salisbury, S.A, Kong Thoo Lin, P, Brown, D.M. | | Deposit date: | 1995-08-01 | | Release date: | 1995-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Direct observation of two base-pairing modes of a cytosine-thymine analogue with guanine in a DNA Z-form duplex: significance for base analogue mutagenesis.
J.Mol.Biol., 251, 1995
|
|
1JE4
 
 | |
228L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1JS7
 
 | | Solution Structure of dAAUAA DNA Bulge | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*UP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
1ZS0
 
 | | Crystal structure of the complex between MMP-8 and a phosphonate inhibitor (S-enantiomer) | | Descriptor: | (1S)-1-{[(4'-METHOXY-1,1'-BIPHENYL-4-YL)SULFONYL]AMINO}-2-METHYLPROPYLPHOSPHONIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Pochetti, G, Gavuzzo, E, Campestre, C, Agamennone, M, Tortorella, P, Consalvi, V, Gallina, C, Hiller, O, Tschesche, H, Tucker, P.A, Mazza, F. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural insight into the stereoselective inhibition of MMP-8 by enantiomeric sulfonamide phosphonates.
J.Med.Chem., 49, 2006
|
|
1ZVO
 
 | | Semi-extended solution structure of human myeloma immunoglobulin D determined by constrained X-ray scattering | | Descriptor: | Immunoglobulin delta heavy chain, myeloma immunoglobulin D lambda | | Authors: | Sun, Z, Almogren, A, Furtado, P.B, Chowdhury, B, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2005-06-02 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-extended Solution Structure of Human Myeloma Immunoglobulin D Determined by Constrained X-ray Scattering.
J.Mol.Biol., 353, 2005
|
|
