6YCY
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Krementsova, E.B, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Full-length Plasmodium falciparum myosin A and essential light chain PfELC structures provide new anti-malarial targets.
Elife, 9, 2020
|
|
8D30
 
 | | Crystal structure of the human COPB2 WD-domains | | Descriptor: | 1,2-ETHANEDIOL, Coatomer subunit beta' | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-31 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human COPB2 WD-domains
To Be Published
|
|
4WML
 
 | | Crystal structure of Saccharomyces cerevisiae OMP synthase in complex with PRP(CH2)P | | Descriptor: | 1-O-[(R)-hydroxy(phosphonomethyl)phosphoryl]-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, Orotate phosphoribosyltransferase 1 | | Authors: | Bang, M.B, Molich, U, Hansen, M.R, Grubmeyer, C, Harris, P. | | Deposit date: | 2014-10-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Saccharomyces cerevisia OMP synthase in complex with PRP(CH2)P
To Be Published
|
|
7NQL
 
 | | 55S mammalian mitochondrial ribosome with ICT1 and P site tRNAMet | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7P35
 
 | | Structure of the SARS-CoV-2 3CL protease in complex with rupintrivir | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Fabrega-Ferrer, M, Perez-Saavedra, J, Herrera-Morande, A, Coll, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
7P36
 
 | | X-ray structure of Lactobacillus kefir alcohol dehydrogenase (wild type) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Transfer of a Rational Crystal Contact Engineering Strategy between Diverse Alcohol Dehydrogenases
Crystals, 11, 2021
|
|
7P7Y
 
 | | X-ray structure of Lactobacillus kefir alcohol dehydrogenase mutant Q126K | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Transfer of a Rational Crystal Contact Engineering Strategy between Diverse Alcohol Dehydrogenases
Crystals, 11, 2021
|
|
8G0U
 
 | |
6NIC
 
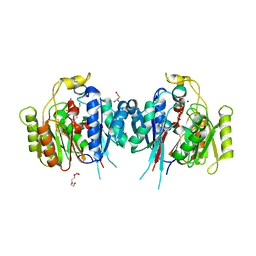 | | Crystal Structure of Medicago truncatula Agmatine Iminohydrolase (Deiminase) in Complex with 6-aminohexanamide | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Study of Agmatine Iminohydrolase FromMedicago truncatula, the Second Enzyme of the Agmatine Route of Putrescine Biosynthesis in Plants.
Front Plant Sci, 10, 2019
|
|
8OI4
 
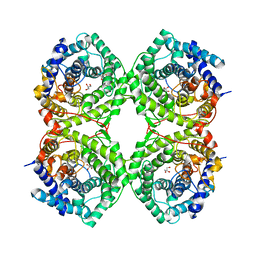 | | Metagenomic Beta-galactosidase from Glycoside Hydrolase family GH154 | | Descriptor: | Beta-galactosidase, CHLORIDE ION, GLYCEROL | | Authors: | Pijning, T, Hameleers, L, Jurak, E, Guskov, A. | | Deposit date: | 2023-03-22 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Novel beta-galactosidase activity and first crystal structure of Glycoside Hydrolase family 154.
N Biotechnol, 80, 2023
|
|
8G0E
 
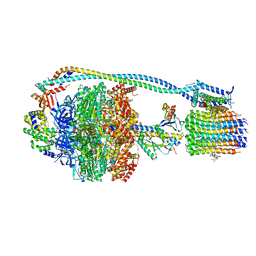 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
6NYV
 
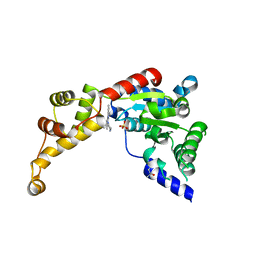 | | Structure of spastin AAA domain in complex with a quinazoline-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, N-(3-tert-butyl-1H-pyrazol-5-yl)-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
8G0D
 
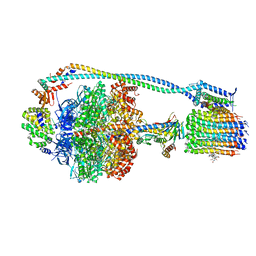 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8W9H
 
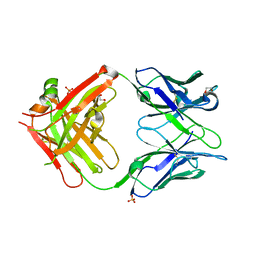 | | Crystal structure of anti-human CLEC12A antibody 50C1 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, anti-human CLEC12A antibody 50C1 Fab Heavy chain, ... | | Authors: | Mori, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2023-09-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of CLEC12A and an antibody that interferes with binding of diverse ligands.
Int.Immunol., 36, 2024
|
|
7OOA
 
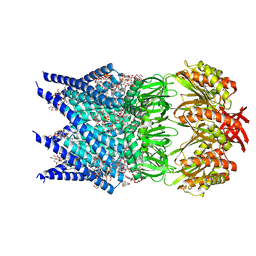 | | Mechanosensitive channel MscS solubilized with LMNG in open conformation with added lipid | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OO0
 
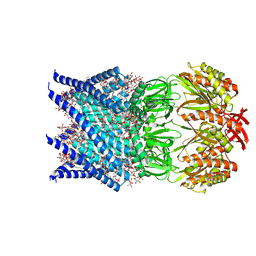 | | Mechanosensitive channel MscS solubilized with DDM in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel MscS | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6NSS
 
 | | TRK-A IN COMPLEX WITH LIGAND 6 | | Descriptor: | High affinity nerve growth factor receptor, N-(8-methyl-2-phenylimidazo[1,2-a]pyrazin-3-yl)-2-(10H-phenoxazin-10-yl)acetamide | | Authors: | Subramanian, G, Brown, D.G. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Deciphering the Allosteric Binding Mechanism of the Human Tropomyosin Receptor Kinase A ( hTrkA) Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
4WV9
 
 | | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane | | Descriptor: | (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J.M, Sankaran, B. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound.
To Be Published
|
|
6ELE
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, fAB heavy chain, ... | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
8TGA
 
 | |
6X0A
 
 | | X-ray structure of a chimeric ParDE toxin-antitoxin complex from Mesorhizobium opportunistum | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Plasmid stabilization system, Putative addiction module antidote protein, ... | | Authors: | Lite, T.L, Grant, R.A, Laub, M.T. | | Deposit date: | 2020-05-15 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library.
Elife, 9, 2020
|
|
6WCT
 
 | |
6O0W
 
 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O2C
 
 | | Crystal structure of 4493 Fab in complex with circumsporozoite protein NANP3 and anti-kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4493 Fab heavy chain, 4493 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
7OMN
 
 | | Anti-EphA1 JD1-1 VH domain | | Descriptor: | GLYCEROL, IMIDAZOLE, JD1-1 VH domain | | Authors: | Ereno Orbea, J, Nilvebrant, J, Sidhu, S, Julien, J.P. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Systematic Engineering of Optimized Autonomous Heavy-Chain Variable Domains.
J.Mol.Biol., 433, 2021
|
|
