3LZI
 
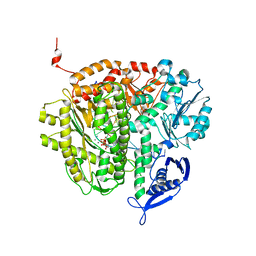 | | RB69 DNA Polymerase (Y567A) ternary complex with dATP Opposite 7,8-dihydro-8-oxoguanine | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-03-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
1HJ7
 
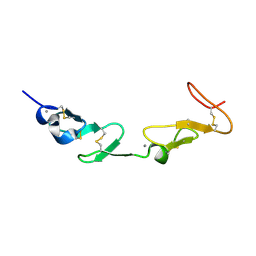 | | NMR study of a pair of LDL receptor Ca2+ binding epidermal growth factor-like domains, 20 structures | | Descriptor: | CALCIUM ION, LDL RECEPTOR | | Authors: | Saha, S, Handford, P.A, Campbell, I.D, Downing, A.K. | | Deposit date: | 2001-01-09 | | Release date: | 2001-07-11 | | Last modified: | 2018-02-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ldl Receptor Egf-Ab Pair: A Paradigm for the Assembly of Tandem Calcium Binding Egf Domains
Structure, 9, 2001
|
|
1FSW
 
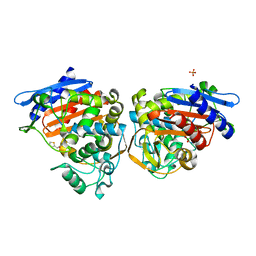 | | AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH INHIBITOR CEPHALOTHINBORONIC ACID | | Descriptor: | CEPHALOSPORINASE, N-2-THIOPHEN-2-YL-ACETAMIDE BORONIC ACID, PHOSPHATE ION | | Authors: | Caselli, E, Powers, R.A, Blaszczak, L.C, Wu, C.Y, Prati, F, Shoichet, B.K. | | Deposit date: | 2000-09-11 | | Release date: | 2001-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Energetic, structural, and antimicrobial analyses of beta-lactam side chain recognition by beta-lactamases.
Chem.Biol., 8, 2001
|
|
3NKU
 
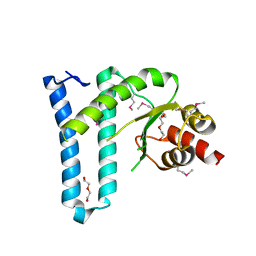 | | Crystal structure of the N-terminal domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, DrrA, TRIETHYLENE GLYCOL | | Authors: | Mueller, M.P, Peters, H, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b.
Science, 329, 2010
|
|
2XEV
 
 | |
1IWJ
 
 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Mutant(109K) Cytochrome P450cam | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
1FSY
 
 | | AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH INHIBITOR CLOXACILLINBORONIC ACID | | Descriptor: | CEPHALOSPORINASE, N-[5-METHYL-3-O-TOLYL-ISOXAZOLE-4-CARBOXYLIC ACID AMIDE] BORONIC ACID, PHOSPHATE ION | | Authors: | Caselli, E, Powers, R.A, Blasczcak, L.C, Wu, C.Y, Prati, F, Shoichet, B.K. | | Deposit date: | 2000-09-11 | | Release date: | 2001-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Energetic, structural, and antimicrobial analyses of beta-lactam side chain recognition by beta-lactamases.
Chem.Biol., 8, 2001
|
|
3EIL
 
 | | Structure of B-DNA d(CGTTAATTAACG)2 in the presence of Manganese | | Descriptor: | 5'-D(*DCP*DGP*DTP*DTP*DAP*DAP*DTP*DTP*DAP*DAP*DCP*DG)-3', MANGANESE (II) ION | | Authors: | Millonig, H, Pous, J, Campos, L.J, Subirana, J.A. | | Deposit date: | 2008-09-16 | | Release date: | 2009-06-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interaction of manganese ions with DNA
J.Inorg.Biochem., 103, 2009
|
|
3LZJ
 
 | | RB69 DNA Polymerase (Y567A) ternary complex with dCTP Opposite 7,8-Dihydro-8-oxoguanine | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-03-01 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
2NW7
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferric heme. Northeast Structural Genomics Target XcR13 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Hussain, A, Bruckmann, C, Thackray, S.J, Seetharaman, J, Tucker, T, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Chapman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4QJV
 
 | |
4QIW
 
 | |
1GE8
 
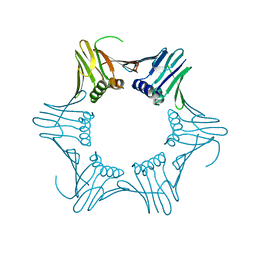 | |
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4RE0
 
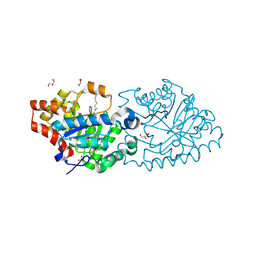 | | Crystal structure of VmoLac in P622 space group | | Descriptor: | COBALT (II) ION, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Hiblot, J, Bzdrenga, J, Champion, C, Gotthard, G, Gonzalez, D, Chabriere, E, Elias, M. | | Deposit date: | 2014-09-20 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia.
Sci Rep, 5, 2015
|
|
4RDZ
 
 | | Crystal structure of VmoLac in P64 space group | | Descriptor: | COBALT (II) ION, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Hiblot, J, Bzdrenga, J, Champion, C, Gotthard, G, Gonzalez, D, Chabriere, E, Elias, M. | | Deposit date: | 2014-09-20 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia.
Sci Rep, 5, 2015
|
|
2F65
 
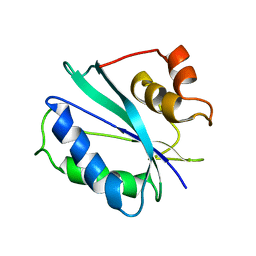 | |
4EQF
 
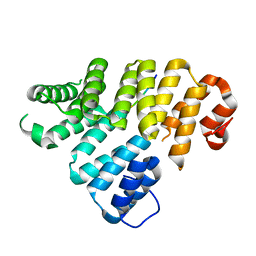 | | Trip8b-1a#206-567 interacting with the carboxy-terminal seven residues of HCN2 | | Descriptor: | PEX5-related protein, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Bankston, J.R, Camp, S.S, Dimaio, F, Lewis, A.S, Chetkovich, D.M, Zagotta, W.N. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ETB
 
 | | lysozyme, room temperature, 200 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ET8
 
 | | Hen egg-white lysozyme solved from 40 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
1DFN
 
 | | CRYSTAL STRUCTURE OF DEFENSIN HNP-3, AN AMPHIPHILIC DIMER: MECHANISMS OF MEMBRANE PERMEABILIZATION | | Descriptor: | DEFENSIN HNP-3 | | Authors: | Hill, C.P, Yee, J, Selsted, M.E, Eisenberg, D. | | Deposit date: | 1991-01-18 | | Release date: | 1992-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization.
Science, 251, 1991
|
|
4ETE
 
 | | Lysozyme, room-temperature, rotating anode, 0.0021 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
2LSC
 
 | | Solution structure of 2'F-ANA and ANA self-complementary duplex | | Descriptor: | DNA (5'-D(*(CFL)P*(GFL)P*(CFL)P*(GFL)P*(A5O)P*(A5O)P*(UAR)P*(UAR)P*(CFL)P*(GFL)P*(CFL)P*(GFL))-3') | | Authors: | Martin-Pintado, N, Yahyaee, M, Campos, R, Noronha, A, Wilds, C, Damha, M, Gonzalez, C. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of double helical arabino nucleic acids (ANA and 2'F-ANA): effect of arabinoses in duplex-hairpin interconversion.
Nucleic Acids Res., 40, 2012
|
|
4ZJF
 
 | | Crystal structure of GP1 - the receptor binding domain of Lassa virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein | | Authors: | Cohen-Dvashi, H, Cohen, N, Israeli, H, Diskin, R. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Molecular Mechanism for LAMP1 Recognition by Lassa Virus.
J.Virol., 89, 2015
|
|
4F2K
 
 | | Macrophage Migration Inhibitory Factor covalently complexed with phenethylisothiocyanate | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, N-(2-phenylethyl)thioformamide, ... | | Authors: | Tyndall, J.D.A, Bernhagen, J, Hampton, M.B, Wilbanks, S.M, Rutledge, M.T. | | Deposit date: | 2012-05-08 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Macrophage migration inhibitory factor covalently complexed with phenethyl isothiocyanate
Acta Crystallogr.,Sect.F, 68, 2012
|
|
