7LE0
 
 | | HIV-1 Protease WT (NL4-3) in Complex with a Darunavir Derivative | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(1,3-benzothiazol-6-yl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
8D8H
 
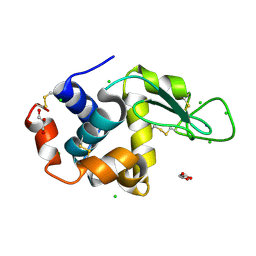 | |
7LDZ
 
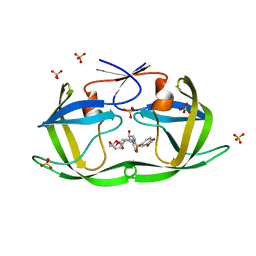 | | HIV-1 Protease WT (NL4-3) in Complex with GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7LE1
 
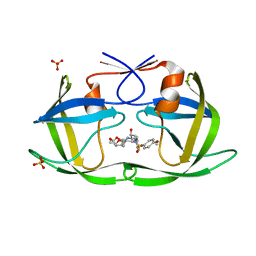 | | HIV-1 Protease WT (NL4-3) in Complex with UMass2 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}propyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7LE2
 
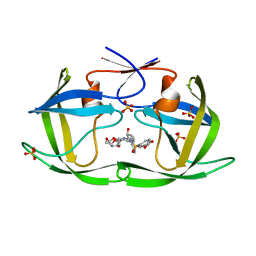 | | HIV-1 Protease WT (NL4-3) in Complex with UMass4 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzodioxol-5-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
6A8D
 
 | | Crystal Structure of Chlamydomonas reinhardtii ARF | | Descriptor: | ARF/SAR superfamily small monomeric GTP binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kumari, S, Goel, M, Kateriya, S, Sharma, P. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of Chlamydomonas reinhardtii Arf
To Be Published
|
|
8D7J
 
 | |
8D1Q
 
 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with 8-Aminoadenosine | | Descriptor: | (2R,3R,4S,5R)-2-(6,8-diaminopurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D22
 
 | |
8D7B
 
 | |
8D5Z
 
 | |
8D7D
 
 | |
7LTD
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 1 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8D8E
 
 | |
6AG7
 
 | | The crystal structure of uPA in complex with HMA-55F | | Descriptor: | 3,5-diamino-N-carbamimidoyl-6-(1-methyl-1H-pyrazol-4-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Majed, H, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | uPA-HMA
To Be Published
|
|
7LTI
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 2 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LTV
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 3 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU0
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 4 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5K06
 
 | | Recombinant bovine beta-lactoglobulin with uncleaved N-terminal methionine (rBlgB) | | Descriptor: | Beta-lactoglobulin, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Polit, A, Hawro, B, Lach, A, Ludwin, E, Lewinski, K. | | Deposit date: | 2016-05-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered beta-Lactoglobulin Produced in E. coli: Purification, Biophysical and Structural Characterisation.
Mol Biotechnol., 58, 2016
|
|
7LU1
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 5 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU2
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 6 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8D5U
 
 | |
7LU3
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 7 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8D6B
 
 | |
7LYX
 
 | | Crystal structure of human CYP8B1 in complex with (S)-tioconazole | | Descriptor: | (S)-Tioconazole, 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase, GLYCEROL, ... | | Authors: | Liu, J, Scott, E.E. | | Deposit date: | 2021-03-08 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and characterization of human cytochrome P450 8B1 supports future drug design for nonalcoholic fatty liver disease and diabetes.
J.Biol.Chem., 298, 2022
|
|
