2I0W
 
 | |
2I0X
 
 | | Hypothetical protein PF1117 from Pyrococcus furiosus | | Descriptor: | Hypothetical protein PF1117 | | Authors: | Chen, L.Q, Fu, Z.-Q, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein Pf1117 from Pyrococcus furiosus
To be Published
|
|
2I0Y
 
 | | cFMS tyrosine kinase (FGF KID) in complex with an arylamide inhibitor | | Descriptor: | 5-CYANO-FURAN-2-CARBOXYLIC ACID [5-HYDROXYMETHYL-2-(4-METHYL-PIPERIDIN-1-YL)-PHENYL]-AMIDE, cFMS Tyrosine Kinase | | Authors: | Schubert, C, Schalk-Hihi, C. | | Deposit date: | 2006-08-11 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of colony-stimulating factor-1 receptor (cFMS) in complex with two inhibitors.
J.Biol.Chem., 282, 2007
|
|
2I0Z
 
 | | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-utilizing dehydrogenases | | Authors: | Minasov, G, Shuvalova, L, Vorontsov, I.I, Kiryukhina, O, Abdullah, J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases
To be Published
|
|
2I10
 
 | | Putative TetR transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | P-NITROPHENOL, Putative TetR transcriptional regulator, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-09-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of putative TetR transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
2I13
 
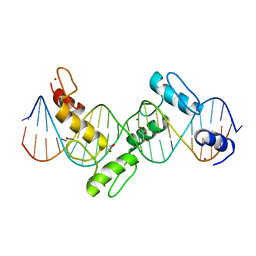 | | Aart, a six finger zinc finger designed to recognize ANN triplets | | Descriptor: | 5'-D(*CP*AP*GP*AP*TP*GP*TP*AP*GP*GP*GP*AP*AP*AP*AP*GP*CP*CP*CP*GP*GP*G)-3', 5'-D(*GP*CP*CP*CP*GP*GP*GP*CP*TP*TP*TP*TP*CP*CP*CP*TP*AP*CP*AP*TP*CP*T)-3', Aart, ... | | Authors: | Horton, N.C, Segal, D.J, Bhakta, M, Crotty, J.W, Barbas III, C.F. | | Deposit date: | 2006-08-12 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of Aart, a Designed Six-finger Zinc Finger Peptide, Bound to DNA.
J.Mol.Biol., 363, 2006
|
|
2I14
 
 | |
2I15
 
 | |
2I16
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594 at temperature of 15K | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Mitshler, A, Hasemann, I, Schneider, T, Cousido, A, Lunin, V.Y, Joachimiak, A, Podjarny, A. | | Deposit date: | 2006-08-13 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Ultrahigh-resolution study of protein atomic displacement parameters at cryotemperatures obtained with a helium cryostat.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I17
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594 at temperature of 60K | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Mitshler, A, Hasemann, I, Schneider, T, Cousido, A, Lunin, V.Y, Joachimiak, A, Podjarny, A. | | Deposit date: | 2006-08-13 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Ultrahigh-resolution study of protein atomic displacement parameters at cryotemperatures obtained with a helium cryostat.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I18
 
 | |
2I19
 
 | | T. Brucei farnesyl diphosphate synthase complexed with bisphosphonate | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [2-(PYRIDIN-2-YLAMINO)ETHANE-1,1-DIYL]BIS(PHOSPHONIC ACID) | | Authors: | Cao, R, Mao, J, Gao, Y, Robinson, H, Odeh, S, Goddard, A, Oldfield, E. | | Deposit date: | 2006-08-13 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Solid-state NMR, crystallographic, and computational investigation of bisphosphonates and farnesyl diphosphate synthase-bisphosphonate complexes.
J.Am.Chem.Soc., 128, 2006
|
|
2I1A
 
 | |
2I1B
 
 | |
2I1D
 
 | | DPC micelle-bound NMR structures of Tritrp1 | | Descriptor: | 13-mer from Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2I1E
 
 | | DPC micelle-bound NMR structures of Tritrp2 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2I1F
 
 | | DPC micelle-bound NMR structures of Tritrp3 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2I1G
 
 | | DPC micelle-bound NMR structures of Tritrp5 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2I1H
 
 | | DPC micelle-bound NMR structures of Tritrp7 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2I1I
 
 | | DPC micelle-bound NMR structures of Tritrp8 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2I1J
 
 | | Moesin from Spodoptera frugiperda at 2.1 angstroms resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Moesin, ... | | Authors: | Li, Q, Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2006-08-14 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Self-masking in an Intact ERM-merlin Protein: An Active Role for the Central alpha-Helical Domain.
J.Mol.Biol., 365, 2007
|
|
2I1K
 
 | |
2I1L
 
 | | Crystal structure of the C2 form of FAD synthetase from Thermotoga maritima | | Descriptor: | Riboflavin kinase/FMN adenylyltransferase | | Authors: | Wang, W, Shin, D.H, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the C2 form of FAD synthetase from Thermotoga maritima
To be Published
|
|
2I1M
 
 | | cFMS tyrosine kinase (tie2 KID) in complex with an arylamide inhibitor | | Descriptor: | 5-CYANO-FURAN-2-CARBOXYLIC ACID [5-HYDROXYMETHYL-2-(4-METHYL-PIPERIDIN-1-YL)-PHENYL]-AMIDE, Macrophage colony-stimulating factor 1 receptor | | Authors: | Schubert, C, Schalk-Hihi, C. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of colony-stimulating factor-1 receptor (cFMS) in complex with two inhibitors.
J.Biol.Chem., 282, 2007
|
|
2I1N
 
 | | Crystal structure of the 1st PDZ domain of Human DLG3 | | Descriptor: | Discs, large homolog 3, SODIUM ION | | Authors: | Turnbull, A.P, Phillips, C, Bunkoczi, G, Debreczeni, J, Ugochukwu, E, Pike, A.C.W, Gorrec, F, Umeano, C, Elkins, J, Berridge, G, Savitsky, P, Gileadi, O, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
