3I01
 
 | | Native structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, water-bound C-cluster. | | Descriptor: | ACETATE ION, COPPER (I) ION, Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, ... | | Authors: | Kung, Y, Doukov, T.I, Drennan, C.L. | | Deposit date: | 2009-06-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
5Z9T
 
 | | a new PL6 alginate lyase complex with trisaccharide | | Descriptor: | GLYCEROL, MALONATE ION, SODIUM ION, ... | | Authors: | Liu, W.Z, Lyu, Q.Q, Zhang, K.K, Li, Z.J. | | Deposit date: | 2018-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into a novel Ca2+-independent PL-6 alginate lyase from Vibrio OU02 identify the possible subsites responsible for product distribution.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
9FK2
 
 | | The structure of glycosynthase XT6 (E265G mutant), the extracellular xylanase of G.proteiniphilus T-6 in complex with two xylobiose-F molecules | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, ZINC ION, ... | | Authors: | Hadad, N, Chmelnik, O, Dessau, M, Shoham, Y, Shoham, G. | | Deposit date: | 2024-06-01 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of glycosynthase XT6 (E265G mutant), the extracellular xylanase of G.proteiniphilus T-6 in complex with two xylobiose-F molecules
To Be Published
|
|
5YSH
 
 | |
1TDR
 
 | | EXPRESSION, CHARACTERIZATION, AND CRYSTALLOGRAPHIC ANALYSIS OF TELLUROMETHIONYL DIHYDROFOLATE REDUCTASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, METHOTREXATE, ... | | Authors: | Lewinski, K, Lebioda, L. | | Deposit date: | 1995-04-13 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Expression, characterization and crystallographic analysis of telluromethionyl dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2WO1
 
 | | Crystal Structure of the EphA4 Ligand Binding Domain | | Descriptor: | EPHRIN TYPE-A RECEPTOR, N-PROPANOL | | Authors: | Bowden, T.A, Aricescu, A.R, Nettleship, J.E, Siebold, C, Rahman-Huq, N, Owens, R.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-07-21 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Plasticity of Eph-Receptor A4 Facilitates Cross-Class Ephrin Signalling
Structure, 17, 2009
|
|
6ML7
 
 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpG 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*(5CM)P*GP*AP*AP*TP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
4H4E
 
 | | IspH in complex with (E)-4-mercapto-3-methylbut-2-enyl diphosphate | | Descriptor: | (2E)-3-methyl-4-sulfanylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Eisenreich, W, Jauch, J, Bacher, A, Groll, M. | | Deposit date: | 2012-09-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Fluoro, Amino, and Thiol Inhibitors Bound to the [Fe(4) S(4) ] Protein IspH.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5W7E
 
 | | Murine acyloxyacyl hydrolase (AOAH), S262A mutant, with dimyristoyl phosphatidylcholine | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acyloxyacyl hydrolase, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the mammalian lipopolysaccharide detoxifier.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3JYQ
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with shikimate and NADH | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Schomburg, D, Niefind, K. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
5U3I
 
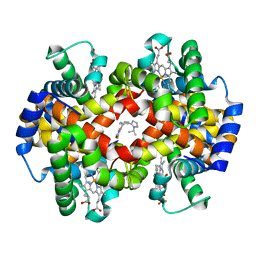 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT compound 31 | | Descriptor: | 2-methoxy-5-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)pyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
5OWK
 
 | |
6FRZ
 
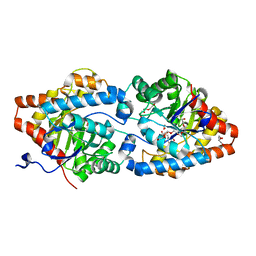 | | Phosphotriesterase PTE_A53_7 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methylcyclohexane-1,1,3,3-tetrol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phosphotriesterase
PTE_A53_7
To Be Published
|
|
3AKB
 
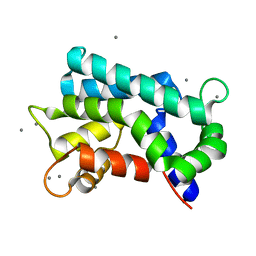 | | Structural basis for prokaryotic calcium-mediated regulation by a Streptomyces coelicolor calcium-binding protein | | Descriptor: | CALCIUM ION, Putative calcium binding protein | | Authors: | Zhao, X, Pang, H, Wang, S, Zhou, W, Yang, K, Bartlam, M. | | Deposit date: | 2010-07-09 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for prokaryotic calciummediated regulation by a Streptomyces coelicolor calcium binding protein
Protein Cell, 1, 2010
|
|
1JKL
 
 | | 1.6A X-RAY STRUCTURE OF BINARY COMPLEX OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE WITH ATP ANALOGUE | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1JKS
 
 | | 1.5A X-RAY STRUCTURE OF APO FORM OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-13 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
5YVK
 
 | | Crystal structure of a cyclase Famc1 from Fischerella ambigua UTEX 1903 | | Descriptor: | CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium, cyclase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.292 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3NQK
 
 | |
7DKM
 
 | | PHGDH covalently linked to oridonin | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, CHLORIDE ION, D-3-phosphoglycerate dehydrogenase, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biophysical and biochemical properties of PHGDH revealed by studies on PHGDH inhibitors.
Cell.Mol.Life Sci., 79, 2021
|
|
7GQD
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z56862798 | | Descriptor: | (1'S)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
5GUW
 
 | | Complex of Cytochrome cd1 Nitrite Reductase and Nitric Oxide Reductase in Denitrification of Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Terasaka, E, Sugimoto, H, Shiro, Y, Tosha, T. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dynamics of nitric oxide controlled by protein complex in bacterial system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7DJM
 
 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.70000112 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
3EA1
 
 | | Crystal Structure of the Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
6M6M
 
 | | The crystal structure of glycosidase mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase | | Authors: | Wang, R.F. | | Deposit date: | 2020-03-15 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization and Structural Elucidation of Enhanced Catalytic Activity upon Engineering Glycosidase KfGH01 for the Production of Vina-ginsenoside R7
To Be Published
|
|
7Z10
 
 | | Monomeric respiratory complex IV isolated from S. cerevisiae | | Descriptor: | COPPER (II) ION, CYTOCHROME C OXIDASE SUBUNIT 3; SYNONYM: CYTOCHROME C OXIDASE POLYPEPTIDE III, COX3, ... | | Authors: | Marechal, A, Hartley, A, Ing, G, Pinotsis, N. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Cryo-EM structure of a monomeric yeast S. cerevisiae complex IV isolated with maltosides: Implications in supercomplex formation.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
