1F0V
 
 | | Crystal structure of an Rnase A dimer displaying a new type of 3D domain swapping | | Descriptor: | 5'-D(*CP*G)-3', GLYCEROL, PHOSPHATE ION, ... | | Authors: | Liu, Y.S, Gotte, G, Libonati, M, Eisenberg, D.S. | | Deposit date: | 2000-05-17 | | Release date: | 2001-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A domain-swapped RNase A dimer with implications for amyloid formation
Nat.Struct.Biol., 8, 2001
|
|
1EZU
 
 | | ECOTIN Y69F, D70P BOUND TO D102N TRYPSIN | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
1B2U
 
 | |
1EZS
 
 | | CRYSTAL STRUCTURE OF ECOTIN MUTANT M84R, W67A, G68A, Y69A, D70A BOUND TO RAT ANIONIC TRYPSIN II | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
1F3X
 
 | | S402P MUTANT OF RABBIT MUSCLE PYRUVATE KINASE | | Descriptor: | MANGANESE (II) ION, POTASSIUM ION, PYRUVATE KINASE, ... | | Authors: | Wooll, J.O, Friesen, R.H.E, White, M.A, Watowich, S.J, Fox, R.O, Lee, J.C, Czerwinski, E.W. | | Deposit date: | 2000-06-06 | | Release date: | 2001-10-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional linkages between subunit interfaces in mammalian pyruvate kinase.
J.Mol.Biol., 312, 2001
|
|
1DP3
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF THE TRAM PROTEIN | | Descriptor: | TRAM PROTEIN | | Authors: | Stockner, T, Plugariu, C, Koraimann, G, Hoegenauer, G, Bermel, W, Prytulla, S, Sterk, H. | | Deposit date: | 1999-12-23 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of TraM.
Biochemistry, 40, 2001
|
|
1F52
 
 | | CRYSTAL STRUCTURE OF GLUTAMINE SYNTHETASE FROM SALMONELLA TYPHIMURIUM CO-CRYSTALLIZED WITH ADP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE SYNTHETASE, ... | | Authors: | Gill, H.S, Pfluegl, G.M.U, Eisenberg, D. | | Deposit date: | 2000-06-12 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition.
Biochemistry, 40, 2001
|
|
1F8A
 
 | | STRUCTURAL BASIS FOR THE PHOSPHOSERINE-PROLINE RECOGNITION BY GROUP IV WW DOMAINS | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1, Y(SEP)PT(SEP)S PEPTIDE | | Authors: | Verdecia, M.A, Bowman, M.E, Lu, K.P, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-29 | | Release date: | 2000-08-23 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for phosphoserine-proline recognition by group IV WW domains.
Nat.Struct.Biol., 7, 2000
|
|
1F8B
 
 | | Native Influenza Virus Neuraminidase in Complex with NEU5AC2EN | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8C
 
 | | Native Influenza Neuraminidase in Complex with 4-amino-2-deoxy-2,3-dehydro-N-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8D
 
 | | Native Influenza Neuraminidase in Complex with 9-amino-2-deoxy-2,3-dehydro-N-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-AMINO-2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8E
 
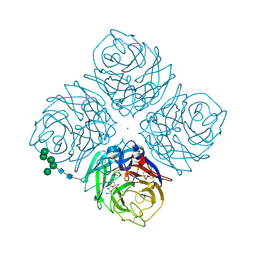 | | Native Influenza Neuraminidase in Complex with 4,9-diamino-2-deoxy-2,3-dehydro-N-acetyl-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,9-AMINO-2,4-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1FEU
 
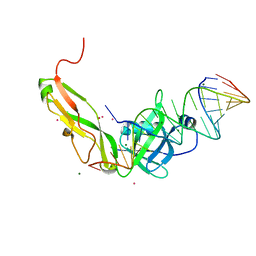 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN TL5, ONE OF THE CTC FAMILY PROTEINS, COMPLEXED WITH A FRAGMENT OF 5S RRNA. | | Descriptor: | 19 NT FRAGMENT OF 5S RRNA, 21 NT FRAGMENT OF 5S RRNA, 50S RIBOSOMAL PROTEIN L25, ... | | Authors: | Fedorov, R.V, Meshcheryakov, V.A, Gongadze, G.M, Fomenkova, N.P, Nevskaya, N.A, Selmer, M, Laurberg, M, Kristensen, O, Al-Karadaghi, S, Liljas, A, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2000-07-23 | | Release date: | 2001-06-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EYV
 
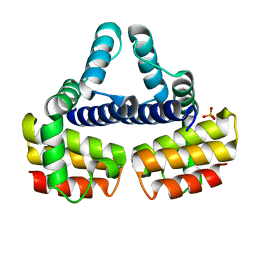 | | THE CRYSTAL STRUCTURE OF NUSB FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | N-UTILIZING SUBSTANCE PROTEIN B HOMOLOG, PHOSPHATE ION | | Authors: | Gopal, B, Haire, L.F, Cox, R.A, Colston, M.J, Major, S, Brannigan, J.A, Smerdon, S.J, Dodson, G.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of NusB from Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1EX4
 
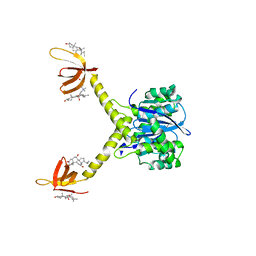 | | HIV-1 INTEGRASE CATALYTIC CORE AND C-TERMINAL DOMAIN | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, INTEGRASE | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EQ2
 
 | | THE CRYSTAL STRUCTURE OF ADP-L-GLYCERO-D-MANNOHEPTOSE 6-EPIMERASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, ADP-L-GLYCERO-D-MANNOHEPTOSE 6-EPIMERASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Deacon, A.M, Ni, Y.S, Coleman Jr, W.G, Ealick, S.E. | | Deposit date: | 2000-03-31 | | Release date: | 2000-11-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of ADP-L-glycero-D-mannoheptose 6-epimerase: catalysis with a twist.
Structure Fold.Des., 8, 2000
|
|
1EUF
 
 | | BOVINE DUODENASE(NEW SERINE PROTEASE), CRYSTAL STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DUODENASE, PHOSPHATE ION | | Authors: | Pletnev, V.Z, Zamolodchikova, T.S, Pangborn, W.A, Duax, W.L. | | Deposit date: | 2000-04-14 | | Release date: | 2001-04-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of bovine duodenase, a serine protease, with dual trypsin and chymotrypsin-like specificities.
Proteins, 41, 2000
|
|
1FR3
 
 | |
1FAY
 
 | | WINGED BEAN ACIDIC LECTIN COMPLEXED WITH METHYL-ALPHA-D-GALACTOSE (MONOCLINIC FORM) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACIDIC LECTIN, CALCIUM ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2000-07-14 | | Release date: | 2001-07-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate specificity and salt-bridge mediated conformational change in acidic winged bean agglutinin.
J.Mol.Biol., 302, 2000
|
|
1FE1
 
 | | CRYSTAL STRUCTURE PHOTOSYSTEM II | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, CADMIUM ION, CHLOROPHYLL A, ... | | Authors: | Zouni, A, Witt, H.-T, Kern, J, Fromme, P, Krauss, N, Saenger, W, Orth, P. | | Deposit date: | 2000-07-20 | | Release date: | 2001-02-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution.
Nature, 409, 2001
|
|
1FBX
 
 | | CRYSTAL STRUCTURE OF ZINC-CONTAINING E.COLI GTP CYCLOHYDROLASE I | | Descriptor: | CHLORIDE ION, GTP CYCLOHYDROLASE I, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, A, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-17 | | Release date: | 2001-02-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FGU
 
 | |
1FJG
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Wimberly, B.T, Morgan-Warren, R.J, Ramakrishnan, V. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics
Nature, 407, 2000
|
|
1FPY
 
 | |
4R72
 
 | | Structure of the periplasmic binding protein AfuA from Actinobacillus pleuropneumoniae (apo form) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC-type Fe3+ transport system, periplasmic component, ... | | Authors: | Sit, B, Calmettes, C, Moraes, T.F. | | Deposit date: | 2014-08-26 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen.
Plos Pathog., 11, 2015
|
|
