3CRV
 
 | | XPD_Helicase | | Descriptor: | CITRATE ANION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
2XHM
 
 | | Crystal structure of AnCE-K26 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ANGIOTENSIN CONVERTING ENZYME, ... | | Authors: | Akif, M, Ntai, I, Sturrock, E.D, Isaac, R.E, Bachmann, B.O, Acharya, K.R. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of a Phosphonotripeptide K-26 in Complex with Angiotensin Converting Enzyme Homologue (Ance) from Drosophila Melanogaster.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
1OO2
 
 | | Crystal structure of transthyretin from Sparus aurata | | Descriptor: | CADMIUM ION, transthyretin | | Authors: | Pasquato, N, Ramazzina, I, Folli, C, Battistutta, R, Berni, R, Zanotti, G. | | Deposit date: | 2003-03-03 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Distinctive binding and structural properties of piscine transthyretin.
Febs Lett., 555, 2003
|
|
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
2EBI
 
 | | Arabidopsis GT-1 DNA-binding domain with T133D phosphomimetic mutation | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Noto, K, Niyada, E, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2007-02-08 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
1L6O
 
 | | XENOPUS DISHEVELLED PDZ DOMAIN | | Descriptor: | Dapper 1, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Cheyette, B.N.R, Waxman, J.S, Miller, J.R, Takemaru, K.-I, Sheldahl, L.C, Khlebtsova, N, Fox, E.P, Earnest, T, Moon, R.T. | | Deposit date: | 2002-03-11 | | Release date: | 2003-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dapper, a Dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation
Dev.Cell, 2, 2002
|
|
2ALG
 
 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | Deposit date: | 2005-08-05 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
1JIG
 
 | | Dlp-2 from Bacillus anthracis | | Descriptor: | Dlp-2, FE (III) ION | | Authors: | Papinutto, E, Dundon, W.G, Pitulis, N, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2001-07-02 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of two iron-binding proteins from Bacillus anthracis.
J.Biol.Chem., 277, 2002
|
|
2FVN
 
 | |
1JI5
 
 | | Dlp-1 from bacillus anthracis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dlp-1, FE (III) ION | | Authors: | Papinutto, E, Dundon, W.G, Pitulis, N, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of two iron-binding proteins from Bacillus anthracis.
J.Biol.Chem., 277, 2002
|
|
1NAE
 
 | | Structure of CsCBM6-3 from Clostridium stercorarium in complex with xylotriose | | Descriptor: | CALCIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, putative xylanase | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G. | | Deposit date: | 2002-11-27 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains
J.Mol.Biol., 327, 2003
|
|
2JMW
 
 | | Structure of DNA-Binding Domain of Arabidopsis GT-1 | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Niyada, E, Noto, K, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2006-12-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
1OD3
 
 | | Structure of CSCBM6-3 From Clostridium stercorarium in complex with laminaribiose | | Descriptor: | ACETIC ACID, CALCIUM ION, PUTATIVE XYLANASE, ... | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
2ESX
 
 | | The structure of the V3 region within gp120 of JR-FL HIV-1 strain (minimized average structure) | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Rosen, O, Sharon, M, Samson, A.O, Quadt, S.R, Anglister, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion.
Proc.Natl.Acad.Sci.Usa, 13, 2006
|
|
2ESZ
 
 | | The structure of the V3 region within gp120 of JR-FL HIV-1 strain (ensemble) | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Rosen, O, Sharon, M, Samson, A.O, Quadt, S.R, Anglister, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion.
Proc.Natl.Acad.Sci.Usa, 13, 2006
|
|
1HA9
 
 | | SOLUTION STRUCTURE OF THE SQUASH TRYPSIN INHIBITOR MCoTI-II, NMR, 30 STRUCTURES. | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Heitz, A, Hernandez, J.-F, Gagnon, J, Hong, T.T, Pham, T.T.C, Nguyen, T.M, Le-Nguyen, D, Chiche, L. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Squash Trypsin Inhibitor Mcoti-II. A New Family for Cyclic Knottins
Biochemistry, 40, 2001
|
|
2KVY
 
 | | NMR solution structure of the 4:1 complex between an uncharged distamycin A analogue and [d(TGGGGT)]4 | | Descriptor: | 4-amino-1-methyl-N-{1-methyl-5-[(1-methyl-5-{[3-(methylamino)-3-oxopropyl]carbamoyl}-1H-pyrrol-3-yl)carbamoyl]-1H-pyrrol-3-yl}-1H-pyrrole-2-carboxamide, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3') | | Authors: | Cosconati, S, Marinelli, L, Trotta, R, Virno, A, De Tito, S, Romagnoli, R, Pagano, B, Limongelli, V, Giancola, C, Baraldi, P, Mayol, L, Novellino, E, Randazzo, A. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle.
J.Am.Chem.Soc., 132, 2010
|
|
2Y68
 
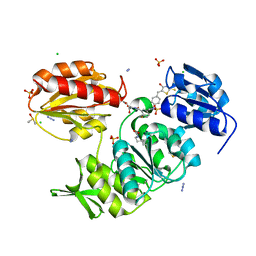 | | Structure-based design of a new series of D-glutamic acid-based inhibitors of bacterial MurD ligase | | Descriptor: | 2-[[2-fluoro-5-[[[4-[(Z)-(4-oxo-2-sulfanylidene-1,3-thiazolidin-5-ylidene)methyl]phenyl]amino]methyl]phenyl]carbonylamino]pentanedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-based design of a new series of D-glutamic acid based inhibitors of bacterial UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase (MurD).
J. Med. Chem., 54, 2011
|
|
1E7K
 
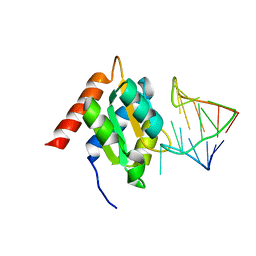 | | Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment | | Descriptor: | 15.5 KD RNA BINDING PROTEIN, RNA (5'-R(*GP*CP*CP*AP*AP*UP*GP*AP*GP*GP*UP*UP*UP* AP*UP*CP*CP*GP*AP*GP*G*C(-3') | | Authors: | Vidovic, I, Nottrott, S, Harthmuth, K, Luhrmann, R, Ficner, R. | | Deposit date: | 2000-08-29 | | Release date: | 2001-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Spliceosomal 15.5Kd Protein Bound to a U4 Snrna Fragment
Mol.Cell, 6, 2000
|
|
1ETE
 
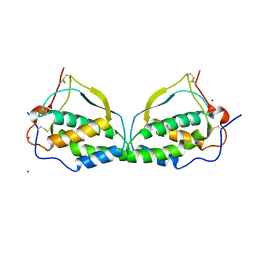 | |
2WJP
 
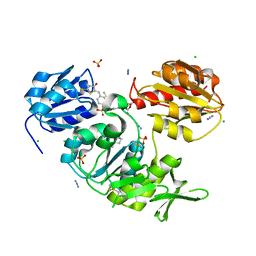 | | CRYSTAL STRUCTURE OF MURD LIGASE IN COMPLEX WITH D-GLU CONTAINING RHODANINE INHIBITOR | | Descriptor: | AZIDE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Rupnik, V, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Muller-Premru, M, Gobec, S, Zega, A, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2009-05-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Novel 5-Benzylidenerhodanine and 5-Benzylidenethiazolidine-2,4-Dione Inhibitors of Murd Ligase.
J.Med.Chem., 53, 2010
|
|
2XGL
 
 | | The X-ray structure of the Escherichia coli colicin M immunity protein demonstrates the presence of a disulphide bridge, which is functionally essential | | Descriptor: | CADMIUM ION, CHLORIDE ION, COLICIN-M IMMUNITY PROTEIN, ... | | Authors: | Gerard, F, Brooks, M.A, Barreteau, H, Touze, T, Graille, M, Bouhss, A, Blanot, D, Tilbeurgh, H.v, Mengin-Lecreulx, D. | | Deposit date: | 2010-06-07 | | Release date: | 2010-11-17 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray Structure and Site-Directed Mutagenesis Analysis of the Escherichia Coli Colicin M Immunity Protein.
J.Bacteriol., 193, 2011
|
|
1BH4
 
 | |
2X5O
 
 | | Discovery of Novel 5-Benzylidenerhodanine- and 5-Benzylidene- thiazolidine-2,4-dione Inhibitors of MurD Ligase | | Descriptor: | AZIDE ION, CHLORIDE ION, N-({3-[({4-[(Z)-(2,4-DIOXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]PHENYL}AMINO)METHYL]PHENYL}CARBONYL)-D-GLUTAMIC ACID, ... | | Authors: | Zidar, N, Tomasic, T, Sink, R, Rupnik, V, Kovac, A, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Gobec, S, Zega, A, Peterlin-Masic, L, Kikelja, D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of novel 5-benzylidenerhodanine and 5-benzylidenethiazolidine-2,4-dione inhibitors of MurD ligase.
J. Med. Chem., 53, 2010
|
|
2Y1O
 
 | | Dual-target Inhibitor of MurD and MurE Ligases: Design, Synthesis and Binding Mode Studies | | Descriptor: | (2R)-2-[[3-[[4-[(Z)-(4-OXO-2-SULFANYLIDENE-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]PHENYL]METHYLAMINO]PHENYL]CARBONYLAMINO]PENTANEDIOIC ACID, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Tomasic, T, Sink, R, Kovac, A, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Gobec, S, Zega, A, Kikelj, D, Peterlin-Masic, L. | | Deposit date: | 2010-12-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Dual Inhibitor of MurD and MurE Ligases from Escherichia coli and Staphylococcus aureus.
ACS Med Chem Lett, 3, 2012
|
|
