2XQ8
 
 | | Pentameric ligand gated ion channel GLIC in complex with zinc ion (Zn2+) | | Descriptor: | GLR4197 PROTEIN, ZINC ION | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQ9
 
 | | Pentameric ligand gated ion channel GLIC mutant E221A in complex with tetraethylarsonium (TEAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
1MNI
 
 | |
1E8C
 
 | | Structure of MurE the UDP-N-acetylmuramyl tripeptide synthetase from E. coli | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CHLORIDE ION, UDP-N-ACETYLMURAMOYLALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Gordon, E.J, Chantala, L, Dideberg, O. | | Deposit date: | 2000-09-19 | | Release date: | 2001-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Udp-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: Meso-Diaminopimelate Ligase from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1B9V
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN TEH ACTIVE SITE | | Descriptor: | 1-[4-CARBOXY-2-(3-PENTYLAMINO)PHENYL]-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zahao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1B9S
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-ACETYLAMINO)-3-[N-(2-ETHYLBUTANOYLAMINO)]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
4M3D
 
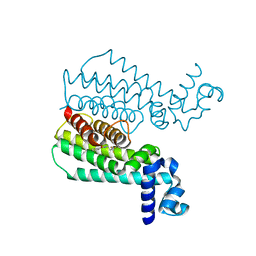 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-{3-[(phenylsulfonyl)amino]prop-1-yn-1-yl}-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4M3E
 
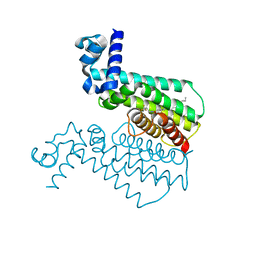 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-{[(propylsulfonyl)amino]methyl}-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
1YT0
 
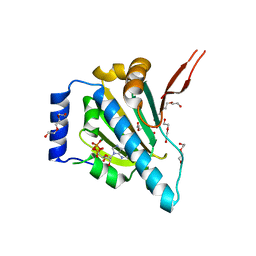 | | Crystal Structure of the Unliganded Form of GRP94, the ER Hsp90: Basis for Nucleotide-Induced Conformational Change, GRP94N(DELTA)41 APO CRYSTAL SOAKED WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Dollins, D.E, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Unliganded GRP94, the Endoplasmic Reticulum Hsp90: BASIS FOR NUCLEOTIDE-INDUCED CONFORMATIONAL CHANGE
J.Biol.Chem., 280, 2005
|
|
4M3G
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzenesulfonamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
3A7U
 
 | |
3GKM
 
 | |
4M3B
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4MH5
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
1EEH
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
1BBH
 
 | |
7V62
 
 | | Crystal structure of human OSBP ORD in complex with cholesterol | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHOLESTEROL, CITRIC ACID, ... | | Authors: | Kobayashi, J, Kato, R. | | Deposit date: | 2021-08-19 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Ligand Recognition by the Lipid Transfer Domain of Human OSBP Is Important for Enterovirus Replication.
Acs Infect Dis., 8, 2022
|
|
2A3A
 
 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with theophylline | | Descriptor: | SULFATE ION, THEOPHYLLINE, chitinase | | Authors: | Rao, F.V, Andersen, O.A, Vora, K.A, DeMartino, J.A, van Aalten, D.M.F. | | Deposit date: | 2005-06-24 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes.
Chem.Biol., 12, 2005
|
|
6RSS
 
 | | Solution structure of the fourth WW domain of WWP2 with GB1-tag | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Wahl, L.C, Watt, J.E, Tolchard, J, Blumenschein, T.M.A, Chantry, A. | | Deposit date: | 2019-05-22 | | Release date: | 2019-10-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Smad7 Binds Differently to Individual and Tandem WW3 and WW4 Domains of WWP2 Ubiquitin Ligase Isoforms.
Int J Mol Sci, 20, 2019
|
|
1H6X
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
1H6Y
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
2A3E
 
 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, SULFATE ION, ... | | Authors: | Rao, F.V, Andersen, O.A, Vora, K.A, DeMartino, J.A, van Aalten, D.M.F. | | Deposit date: | 2005-06-24 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes.
Chem.Biol., 12, 2005
|
|
2A3B
 
 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with caffeine | | Descriptor: | CAFFEINE, SULFATE ION, chitinase | | Authors: | Rao, F.V, Andersen, O.A, Vora, K.A, DeMartino, J.A, van Aalten, D.M.F. | | Deposit date: | 2005-06-24 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes.
Chem.Biol., 12, 2005
|
|
2R2X
 
 | | Ricin A-chain (recombinant) complex with Urea | | Descriptor: | Ricin A chain, SULFATE ION, UREA | | Authors: | Carra, J.H, McHugh, C.A, Mulligan, S, Machiesky, L.M, Soares, A.S, Millard, C.B. | | Deposit date: | 2007-08-28 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based identification of determinants of conformational and spectroscopic change at the ricin active site.
Bmc Struct.Biol., 7, 2007
|
|
7Q43
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10 | | Descriptor: | CITRIC ACID, Dedicator of cytokinesis protein 10 peptide, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40002346 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10
To Be Published
|
|
