8U64
 
 | | Cryo-EM structure of PsBphP in Pfr state, medial PSM only | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
9MHB
 
 | | Crystal Structure of Human P38 alpha MAPK In Complex with MW01-14-064SRM | | Descriptor: | (5P)-N,N-dimethyl-5-(naphthalen-1-yl)-6-(pyridin-4-yl)pyrazin-2-amine, 1,2-ETHANEDIOL, 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, ... | | Authors: | Minasov, G, Tokars, V.L, Roy, S.M, Watterson, D.M, Shuvalova, L. | | Deposit date: | 2024-12-11 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Human P38 alpha MAPK In Complex with MW01-14-064SRM
To Be Published
|
|
8U8Z
 
 | | Cryo-EM structure of PsBphP in Pr state, extended DHp | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
3JXQ
 
 | | X-Ray structure of r[CGCG(5-fluoro)CG]2 | | Descriptor: | MAGNESIUM ION, r[CGCG(5-fluoro)CG]2 | | Authors: | Adamiak, D.A, Milecki, J, Adamiak, R.W, Rypniewski, W. | | Deposit date: | 2009-09-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The hydration and unusual hydrogen bonding in the crystal structure of an RNA duplex containing alternating CG base pairs
New J.Chem., 34, 2010
|
|
8ATX
 
 | |
5IPG
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure FFT-butyl (Hyperoxodized by t-butyl hydroperoxide) | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
6H6N
 
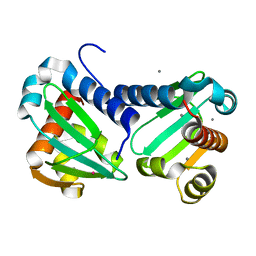 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | CALCIUM ION, TERBIUM(III) ION, Ubiquinone biosynthesis protein UbiJ | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6QN9
 
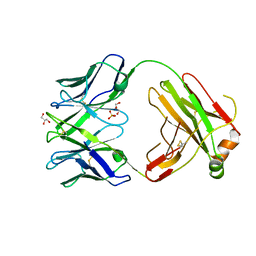 | | Structure of bovine anti-RSV Fab B4 | | Descriptor: | GLYCEROL, Heavy chain, SULFATE ION, ... | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
7NT6
 
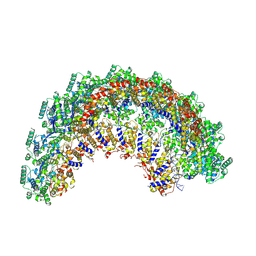 | | CryoEM structure of the Nipah virus nucleocapsid spiral clam-shaped assembly | | Descriptor: | Nucleoprotein, RNA (42-MER), RNA (48-MER) | | Authors: | Ker, D.S, Jenkins, H.T, Greive, S.J, Antson, A.A. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM structure of the Nipah virus nucleocapsid assembly.
Plos Pathog., 17, 2021
|
|
8U4X
 
 | | Cryo-EM structure of PsBphP in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
8U62
 
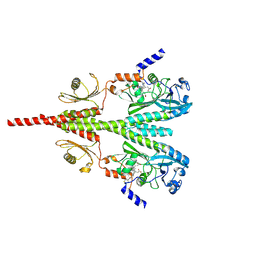 | | Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers FL | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, histidine kinase | | Authors: | Basore, K, Burgie, E.S, Vierstra, D. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.
Nat Commun, 15, 2024
|
|
6TFD
 
 | | Crystal structure of nitrite and NO bound three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
6H7B
 
 | | Structure of Leishmania PABP1 (domain J) complexed with a peptide containing the PAM2 motif of eIF4E4. | | Descriptor: | HIS-HIS-MET-ASN-PRO-ASN-ALA-THR-GLU-PHE-MET-PRO, Polyadenylate-binding protein | | Authors: | Cameron, A.D, Firczuk, H, dos Santos Rodrigues, F.H, McCarthy, J.E.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Leishmania PABP1-eIF4E4 interface: a novel 5'-3' interaction architecture for trans-spliced mRNAs.
Nucleic Acids Res., 47, 2019
|
|
8IHR
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | Descriptor: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
7UHT
 
 | | SSX Structure of Metallo Beta-Lactamase L1 with One Zinc in the Active Site | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | Descriptor: | Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
4AR7
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2012-04-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structure of a Fluorescent Protein from Aequorea Victoria Bearing the Obligate-Monomer Mutation A206K.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1G7K
 
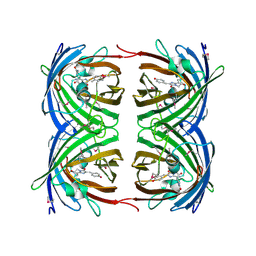 | | CRYSTAL STRUCTURE OF DSRED, A RED FLUORESCENT PROTEIN FROM DISCOSOMA SP. RED | | Descriptor: | FLUORESCENT PROTEIN FP583 | | Authors: | Yarbrough, D, Wachter, R.M, Kallio, K, Matz, M.V, Remington, S.J. | | Deposit date: | 2000-11-10 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7NT5
 
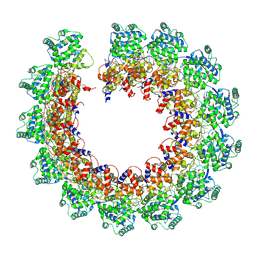 | | CryoEM structure of the Nipah virus nucleocapsid single helical turn assembly | | Descriptor: | Nucleoprotein, RNA (78-MER) | | Authors: | Ker, D.S, Jenkins, H.T, Greive, S.J, Antson, A.A. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM structure of the Nipah virus nucleocapsid assembly.
Plos Pathog., 17, 2021
|
|
5J4C
 
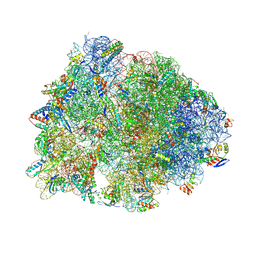 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with cisplatin (soaked) and bound to mRNA and A-, P- and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Melnikov, S.V, Soll, D, Steitz, T.A, Polikanov, Y.S. | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-27 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into RNA binding by the anticancer drug cisplatin from the crystal structure of cisplatin-modified ribosome.
Nucleic Acids Res., 44, 2016
|
|
8ATU
 
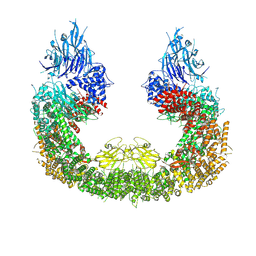 | | Cryo-EM structure of human BIRC6 | | Descriptor: | Baculoviral IAP repeat-containing protein 6, ZINC ION | | Authors: | Ehrmann, J.F, Grabarczyk, D.B, Clausen, T. | | Deposit date: | 2022-08-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex.
Science, 379, 2023
|
|
7O9J
 
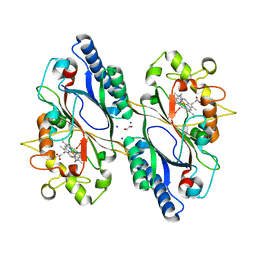 | | Crystal structure of DyP-type peroxidase from Dictyostelium discoideum in complex with an activated form of oxygen | | Descriptor: | 1,2-ETHANEDIOL, DyPA, OXYGEN MOLECULE, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
7UHR
 
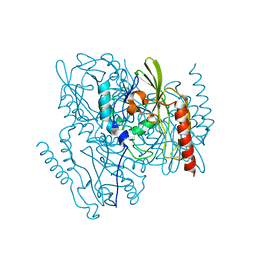 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 Before Reaction (Dark-Set) | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
6OE2
 
 | |
