3CWT
 
 | | Crystal Structure of an AlkA Host/Guest Complex 2'-fluoro-2'-deoxyinosine:Adenine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(2FI)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DAP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3D38
 
 | | Crystal structure of new trigonal form of photosynthetic reaction center from Blastochloris viridis. Crystals grown in microfluidics by detergent capture. | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Nachtergaele, S.H.M, Seddon, A.M, Tereshko, V, Ponomarenko, N, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Simple host-guest chemistry to modulate the process of concentration and crystallization of membrane proteins by detergent capture in a microfluidic device.
J.Am.Chem.Soc., 130, 2008
|
|
3E7N
 
 | | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2 | | Descriptor: | 1,2-ETHANEDIOL, D-ribose high-affinity transport system | | Authors: | Nocek, B, Maltseva, N, Gu, M, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-18 | | Release date: | 2008-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2
To be Published
|
|
3E8F
 
 | |
3DTU
 
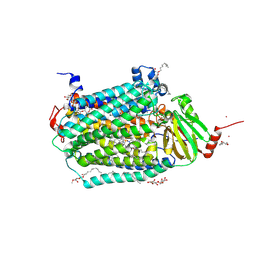 | | Catalytic core subunits (I and II) of cytochrome c oxidase from Rhodobacter sphaeroides complexed with deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CADMIUM ION, CALCIUM ION, ... | | Authors: | Qin, L, Mills, D.A, Buhrow, L, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A conserved steroid binding site in cytochrome C oxidase.
Biochemistry, 47, 2008
|
|
3DUG
 
 | | Crystal structure of zn-dependent arginine carboxypeptidase complexed with zinc | | Descriptor: | ARGININE, GLYCEROL, ZINC ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
3E08
 
 | | H55S mutant Xanthomonas campestris tryptophan 2,3-dioxygenase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, Tryptophan 2,3-dioxygenase | | Authors: | Mowat, C.G, Campbell, L.P. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling substrate binding
Biochemistry, 47, 2008
|
|
3E1Q
 
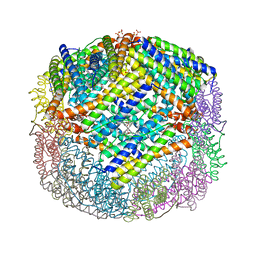 | | Crystal structure of W133F variant E. coli Bacterioferritn with iron. | | Descriptor: | BACTERIOFERRITIN, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crow, A, Lawson, T.L, Lewin, A, Moore, G.R, Le Brun, N.E. | | Deposit date: | 2008-08-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Monitoring the iron status of the ferroxidase center of Escherichia coli bacterioferritin using fluorescence spectroscopy.
Biochemistry, 48, 2009
|
|
3E1N
 
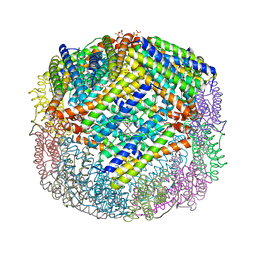 | | Crystal structure of E. coli Bacterioferritin (BFR) after a 65 minute (aerobic) exposure to FE(II) revealing a possible MU-OXO bridge/MU-Hydroxy bridged DIIRON intermediate at the ferroxidase centre. (FE(III)-O-FE(III)-BFR). | | Descriptor: | BACTERIOFERRITIN, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
3BVF
 
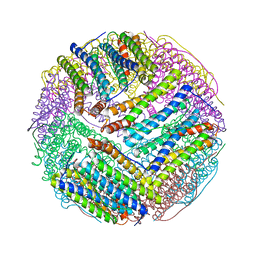 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
3E8B
 
 | |
3EKE
 
 | | Crystal structure of IBV X-domain at pH 5.6 | | Descriptor: | L(+)-TARTARIC ACID, Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
3CWU
 
 | | Crystal Structure of an AlkA Host/Guest Complex 2'-fluoro-2'-deoxy-1,N6-ethenoadenine:Thymine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(2FE)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DTP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CUR
 
 | | Structure of a double methionine mutant of NI-FE hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A. | | Deposit date: | 2008-04-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental approaches to kinetics of gas diffusion in hydrogenase
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CWS
 
 | | Crystal Structure of an AlkA Host/Guest Complex 2'-fluoro-2'-deoxyinosine:Thymine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(2FI)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DTP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3D64
 
 | |
3CVO
 
 | |
3CWA
 
 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Cytosine Base Pair | | Descriptor: | DNA (5'-D(*(8OG)P*DAP*DCP*DAP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3D4V
 
 | | Crystal Structure of an AlkA Host/Guest Complex N7MethylGuanine:Cytosine Base Pair | | Descriptor: | 5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(FMG)P*DTP*DGP*DCP*DC)-3', 5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Lee, S, Bowman, B.R, Wang, S, Verdine, G.L. | | Deposit date: | 2008-05-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine.
J.Am.Chem.Soc., 130, 2008
|
|
3CX5
 
 | |
3E3I
 
 | |
3EF2
 
 | | Structure of the Marasmius oreades mushroom lectin (MOA) in complex with Galalpha(1,3)[Fucalpha(1,2)]Gal and Calcium. | | Descriptor: | ACETATE ION, Agglutinin, CALCIUM ION, ... | | Authors: | Grahn, E.M, Goldstein, I.J, Krengel, U. | | Deposit date: | 2008-09-08 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of a Lectin from the Mushroom Marasmius oreades in Complex with the Blood Group B Trisaccharide and Calcium.
J.Mol.Biol., 390, 2009
|
|
3C3P
 
 | |
3C5Y
 
 | |
3D1N
 
 | | Structure of human Brn-5 transcription factor in complex with corticotrophin-releasing hormone gene promoter | | Descriptor: | 5'-D(*DAP*DGP*DCP*DAP*DTP*DAP*DAP*DAP*DTP*DAP*DAP*DTP*DAP*DA)-3', 5'-D(*DTP*DTP*DAP*DTP*DTP*DAP*DTP*DTP*DTP*DAP*DTP*DGP*DCP*DT)-3', POU domain, ... | | Authors: | Pereira, J.H, Ha, S.C, Kim, S.-H. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of human Brn-5 transcription factor in complex with CRH gene promoter.
J.Struct.Biol., 167, 2009
|
|
