2KG9
 
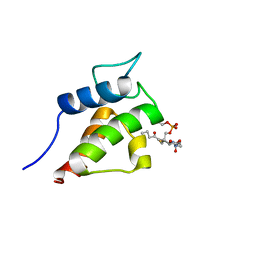 | | NMR Solution Structures of butyryl-ACP (a non-polar, non pathway intermediate) from the actinorhodin polyketide synthase in Streptomyces coelicolor | | Descriptor: | Actinorhodin polyketide synthase acyl carrier protein, THIOBUTYRIC ACID S-{2-[3-(2-HYDROXY-3,3-DIMETHYL-4-PHOSPHONOOXY-BUTYRYLAMINO)-PROPIONYLAMINO]-ETHYL} ESTER | | Authors: | Crump, M.P, Evans, S.E, Williams, C. | | Deposit date: | 2009-03-07 | | Release date: | 2009-04-14 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Probing the Interactions of Early Polyketide Intermediates with the Actinorhodin ACP from S. coelicolor A3(2).
J.Mol.Biol., 389, 2009
|
|
2KG8
 
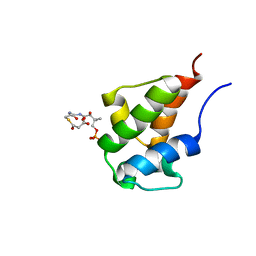 | | NMR Solution Structures of malonyl ACP from the actinorhodin polyketide synthase in Streptomyces coelicolor | | Descriptor: | 3-{[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-3-oxopropanoic acid, Actinorhodin polyketide synthase acyl carrier protein | | Authors: | Crump, M.P, Evans, S.E, Williams, C, Eliza, P. | | Deposit date: | 2009-03-06 | | Release date: | 2009-04-14 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Probing the Interactions of Early Polyketide Intermediates with the Actinorhodin ACP from S. coelicolor A3(2).
J.Mol.Biol., 389, 2009
|
|
2KGA
 
 | |
2KDL
 
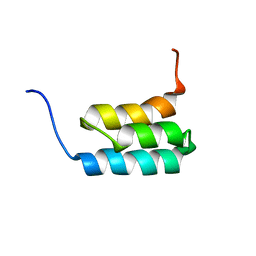 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | Descriptor: | designed protein | | Authors: | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KOS
 
 | |
2KOR
 
 | |
2KOQ
 
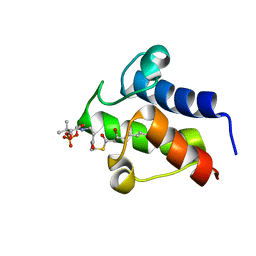 | | NMR solution structures of 3-hydroxyoctanoyl-ACP from Streptomyces coelicolor Fatty Acid Synthase | | Descriptor: | Acyl carrier protein, S-[2-[3-[[(2S)-2-hydroxy-3,3-dimethyl-4-phosphonooxy-butanoyl]amino]propanoylamino]ethyl] (3R)-3-hydroxyoctanethioate | | Authors: | Ploskon, E, Arthur, C.J, Kanari, A.J, Crump, M.P. | | Deposit date: | 2009-09-29 | | Release date: | 2010-08-18 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Recognition of intermediate functionality by acyl carrier protein over a complete cycle of fatty acid biosynthesis
Chem.Biol., 17, 2010
|
|
1RCS
 
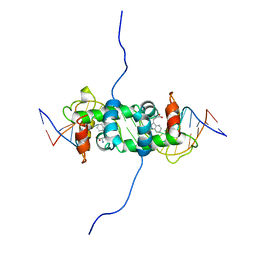 | | NMR STUDY OF TRP REPRESSOR-OPERATOR DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*AP*CP*G)-3'), TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the trp repressor-operator DNA complex.
J.Mol.Biol., 238, 1994
|
|
2K1Q
 
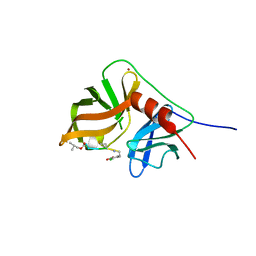 | | NMR structure of hepatitis c virus ns3 serine protease complexed with the non-covalently bound phenethylamide inhibitor | | Descriptor: | NS3 PROTEASE, PHENETHYLAMIDE, ZINC ION | | Authors: | Eliseo, T, Gallo, M, Pennestri, M, Bazzo, R, Cicero, D.O. | | Deposit date: | 2008-03-13 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Binding of a noncovalent inhibitor exploiting the S' region stabilizes the hepatitis C virus NS3 protease conformation in the absence of cofactor.
J.Mol.Biol., 385, 2009
|
|
1K0T
 
 | | NMR SOLUTION STRUCTURE OF UNBOUND, OXIDIZED PHOTOSYSTEM I SUBUNIT PSAC, CONTAINING [4FE-4S] CLUSTERS FA AND FB | | Descriptor: | IRON/SULFUR CLUSTER, PSAC SUBUNIT OF PHOTOSYSTEM I | | Authors: | Antonkine, M.L, Liu, G, Bentrop, D, Bryant, D.A, Bertini, I, Luchinat, C, Golbeck, J.H, Stehlik, D. | | Deposit date: | 2001-09-20 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the unbound, oxidized Photosystem I subunit PsaC, containing [4Fe-4S] clusters F(A) and F(B): a conformational change occurs upon binding to photosystem I.
J.Biol.Inorg.Chem., 7, 2002
|
|
1G5E
 
 | |
1GJ0
 
 | |
1G5D
 
 | |
1GIZ
 
 | |
1RQT
 
 | | NMR structure of dimeric N-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
2RRS
 
 | |
2GVS
 
 | | NMR solution structure of CSPsg4 | | Descriptor: | chemosensory protein CSP-sg4 | | Authors: | Tomaselli, S, Crescenzi, O, Sanfelice, D, Ab, E, Tancredi, T, Picone, D. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Chemosensory Protein from the Desert Locust Schistocerca gregaria(,).
Biochemistry, 45, 2006
|
|
2HW0
 
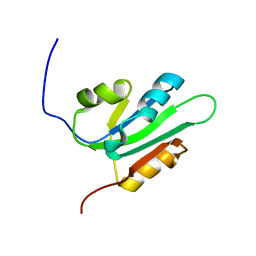 | | NMR Solution Structure of the nuclease domain from the Replicator Initiator Protein from porcine circovirus PCV2 | | Descriptor: | Replicase | | Authors: | Vega-Rocha, S, Byeon, I.L, Gronenborn, B, Gronenborn, A.M, Campos-Olivas, R. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2
J.Mol.Biol., 367, 2007
|
|
1K8B
 
 | |
1K81
 
 | |
1K8J
 
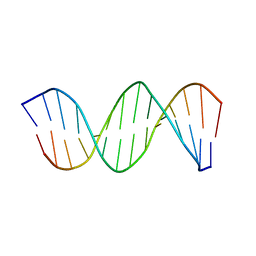 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1KMA
 
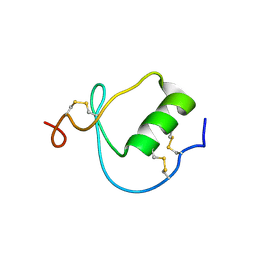 | | NMR Structure of the Domain-I of the Kazal-type Thrombin Inhibitor Dipetalin | | Descriptor: | DIPETALIN | | Authors: | Schlott, B, Wohnert, J, Icke, C, Hartmann, M, Ramachandran, R, Guhrs, K.-H, Glusa, E, Flemming, J, Gorlach, M, Grosse, F, Ohlenschlager, O. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Interaction of Kazal-type inhibitor domains with serine proteinases: biochemical and structural studies.
J.Mol.Biol., 318, 2002
|
|
1N0O
 
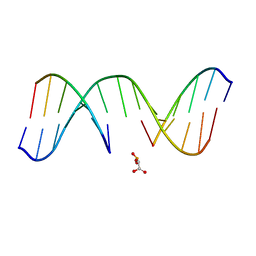 | | NMR Structure of d(CCAAGGXCTTGGG), X is a 3'-phosphoglycolate, 5'-phosphate gapped lesion, 10 structures | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*GP*G)-3', 5'-D(*CP*CP*CP*AP*AP*GP*GP*CP*CP*TP*TP*GP*G)-3', ... | | Authors: | Junker, H.-D, Hoehn, S.T, Bunt, R.C, Marathius, V, Chen, J, Turner, C.J, Stubbe, J. | | Deposit date: | 2002-10-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Characterization, and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Nucleic Acids Res., 30, 2002
|
|
1N0K
 
 | | NMR Structure of duplex DNA d(CCAAGGXCTTGGG), X is a 3' phosphoglycolate, 5'phosphate gapped lesion | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*GP*G)-3', 5'-D(*CP*CP*CP*AP*AP*GP*GP*CP*CP*TP*TP*GP*G)-3', ... | | Authors: | Junker, H.-D, Hoehn, S.T, Bunt, R.C, Marathius, V, Chen, J, Turner, C.J, Stubbe, J. | | Deposit date: | 2002-10-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Characterization and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Nucleic Acids Res., 30, 2002
|
|
