2BPP
 
 | |
2BPQ
 
 | | Anthranilate phosphoribosyltransferase (TrpD) from Mycobacterium tuberculosis (Apo structure) | | Descriptor: | ANTHRANILATE PHOSPHORIBOSYLTRANSFERASE, BENZAMIDINE, GLYCEROL | | Authors: | Lee, C.E, Goodfellow, C, Javid-Majd, F, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-04-22 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Trpd, a Metabolic Enzyme Essential for Lung Colonization by Mycobacterium Tuberculosis, in Complex with its Substrate Phosphoribosylpyrophosphate
J.Mol.Biol., 355, 2006
|
|
2BPR
 
 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, 25 STRUCTURES | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
2BPS
 
 | |
2BPU
 
 | | The Kedge Holmium Derivative of Hen Egg-White Lysozyme at high resolution from Single Wavelength Anomalous Diffraction | | Descriptor: | CHLORIDE ION, HOLMIUM ATOM, LYSOZYME C, ... | | Authors: | Jakoncic, J, Di Michiel, M, Zhong, Z, Honkimaki, V, Jouanneau, Y, Stojanoff, V. | | Deposit date: | 2005-04-25 | | Release date: | 2006-08-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Anomalous Diffraction at Ultra-High Energy for Protein Crystallography.
J.Appl.Crystallogr., 39, 2006
|
|
2BPV
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | 1-[2-HYDROXY-4-(2-HYDROXY-5-METHYL-CYCLOPENTYLCARBAMOYL)5-PHENYL-PENTYL]-4-(3-PYRIDIN-3-YL-PROPIONYL)-PIPERAZINE-2-CARB OXYLIC ACID TERT-BUTYLAMIDE, HIV-1 PROTEASE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPW
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | 1-[2-HYDROXY-4-(2-HYDROXY-5-METHYL-CYCLOPENTYLCARBAMOYL)5-PHENYL-PENTYL]-4-(3-PYRIDIN-3-YL-PROPIONYL)-PIPERAZINE-2-CARB OXYLIC ACID TERT-BUTYLAMIDE, HIV-1 PROTEASE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPX
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPY
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(S)-CYCLOPENTYL-1(R)-HYDROXY-3(R)METHYL]-5-[(2(S)-TERTIARY-BUTYLAMINO-CARBONYL)-4-(N1-(2)-(N-METHYLPIPERAZINYL)-3-CHLORO-PYRAZINYL-5-CARBONYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYL-PENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPZ
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(S)-CYCLOPENTYL-1(R)-HYDROXY-3(R)METHYL]-5-[(2(S)-TERTIARY-BUTYLAMINO-CARBONYL)-4-(N1-(2)-(N-METHYLPIPERAZINYL)-3-CHLORO-PYRAZINYL-5-CARBONYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYL-PENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BQ0
 
 | | 14-3-3 Protein Beta (Human) | | Descriptor: | 14-3-3 BETA/ALPHA | | Authors: | Yang, X, Elkins, J.M, Fedorov, O, Longman, E.J, Sobott, L, Ball, L.J, Sundstrom, M, Arrowsmith, C, Edwards, A, Doyle, D.A. | | Deposit date: | 2005-04-26 | | Release date: | 2005-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2BQ1
 
 | | Ribonucleotide reductase class 1b holocomplex R1E,R2F from Salmonella typhimurium | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Uppsten, M, Farnegardh, M, Domkin, V, Uhlin, U. | | Deposit date: | 2005-04-26 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | The First Holocomplex Structure of Ribonucleotide Reductase Gives New Insight Into its Mechanism of Action
J.Mol.Biol., 359, 2006
|
|
2BQ2
 
 | | Solution Structure of the DNA Duplex ACGCGU-NA with a 2' Amido-Linked Nalidixic Acid Residue at the 3' Terminal Nucleotide | | Descriptor: | 5'-D(*AP*CP*GP*CP*GP*2AU)-3', NALIDIXIC ACID | | Authors: | Siegmund, K, Maheshwary, S, Narayanan, S, Connors, W, Richert, M. | | Deposit date: | 2005-04-26 | | Release date: | 2006-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular details of quinolone-DNA interactions: solution structure of an unusually stable DNA duplex with covalently linked nalidixic acid residues and non-covalent complexes derived from it.
Nucleic Acids Res., 33, 2005
|
|
2BQ3
 
 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*A)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', CALCIUM ION, ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
2BQ4
 
 | | Crystal structure of type I cytochrome c3 from Desulfovibrio africanus | | Descriptor: | BASIC CYTOCHROME C3, CALCIUM ION, HEME C | | Authors: | Czjzek, M, Pieulle, L, Morelli, X, Guerlesquin, F, Hatchikian, E.C. | | Deposit date: | 2005-04-27 | | Release date: | 2005-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Type I / Type II Cytochrome C(3) Complex: An Electron Transfer Link in the Hydrogen-Sulfate Reduction Pathway.
J.Mol.Biol., 354, 2005
|
|
2BQ5
 
 | | MS2 (N87AE89K mutant) - RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-04-27 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BQ6
 
 | | Crystal structure of factor Xa in complex with 21 | | Descriptor: | 1-{[5-(5-CHLORO-2-THIENYL)ISOXAZOL-3-YL]METHYL}-3-CYANO-N-(1-ISOPROPYLPIPERIDIN-4-YL)-7-METHYL-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BQ7
 
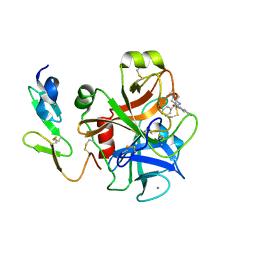 | | Crystal structure of factor Xa in complex with 43 | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, FACTOR XA, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BQ8
 
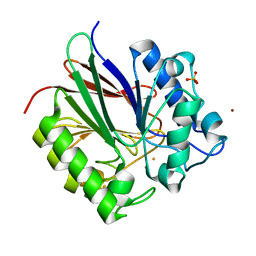 | | Crystal structure of human purple acid phosphatase with an inhibitory conformation of the repression loop | | Descriptor: | FE (II) ION, SULFATE ION, TARTRATE-RESISTANT ACID PHOSPHATASE TYPE 5, ... | | Authors: | Straeter, N, Jasper, B, Krebs, B. | | Deposit date: | 2005-04-27 | | Release date: | 2005-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Recombinant Human Purple Acid Phosphatase with and without an Inhibitory Conformation of the Repression Loop.
J.Mol.Biol., 351, 2005
|
|
2BQA
 
 | |
2BQB
 
 | |
2BQC
 
 | |
2BQD
 
 | |
2BQE
 
 | |
2BQF
 
 | |
