7AE7
 
 | |
7JMD
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7AE4
 
 | |
8OPC
 
 | |
7SLF
 
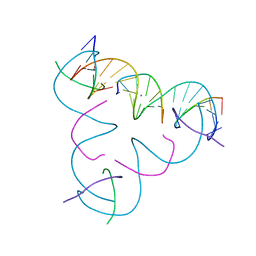 | | [T:Ag+:T--pH 10.5] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.83 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
7AAQ
 
 | | sugar/H+ symporter STP10 in outward occluded conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ACETATE ION, ... | | Authors: | Bavnhoej, L, Paulsen, P.A, Pedersen, B.P. | | Deposit date: | 2020-09-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Molecular mechanism of sugar transport in plants unveiled by structures of glucose/H + symporter STP10.
Nat.Plants, 7, 2021
|
|
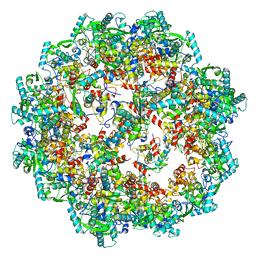
8OPJ
 
 | |
7S9J
 
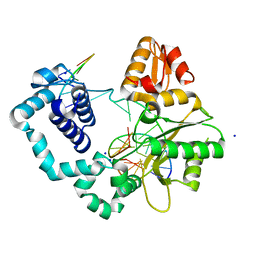 | | Crystal Structure of DNA Polymerase Beta with Fapy-dG base-paired with a dC | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*(FAP)P*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Ryan, B.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.
J.Am.Chem.Soc., 144, 2022
|
|
2YDG
 
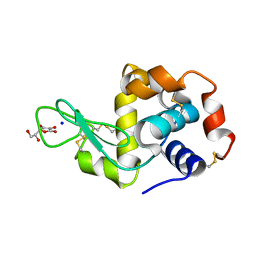 | | Ascorbate co-crystallized HEWL. | | Descriptor: | ASCORBIC ACID, Lysozyme C, SODIUM ION | | Authors: | De la Mora, E, Carmichael, I, Garman, E.F. | | Deposit date: | 2011-03-19 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effective Scavenging at Cryotemperatures: Further Increasing the Dose Tolerance of Protein Crystals.
J.Synchrotron.Radiat., 18, 2011
|
|
7PG4
 
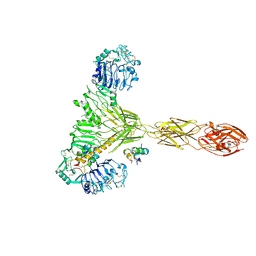 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 2 insulin, conf 3 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
8OOX
 
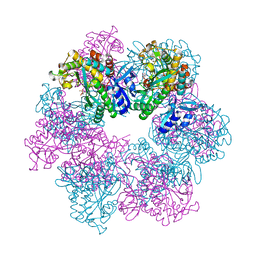 | | Glutamine synthetase from Methermicoccus shengliensis at a resolution of 3.09 A | | Descriptor: | CITRIC ACID, GLYCEROL, Glutamine synthetase, ... | | Authors: | Mueller, M.-C, Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Differences in regulation mechanisms of glutamine synthetases from methanogenic archaea unveiled by structural investigations.
Commun Biol, 7, 2024
|
|
7DOI
 
 | |
3H9E
 
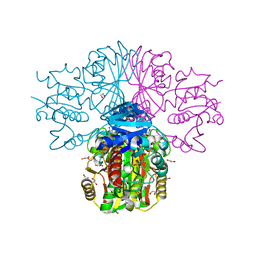 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD and phosphate | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
8OPG
 
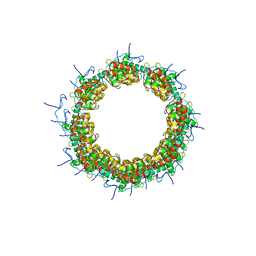 | |
7JQ2
 
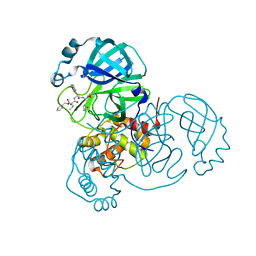 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI5 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
2Y9T
 
 | |
2Y95
 
 | |
7S9P
 
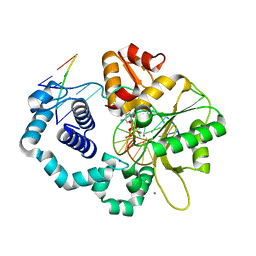 | | Ternary complex of DNA Polymerase Beta with Template Fapy-dG and an incoming dCTP analog | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*(FAP)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Ryan, B.J, Smith, M.R. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.
J.Am.Chem.Soc., 144, 2022
|
|
7PV9
 
 | |
7PDB
 
 | |
2M57
 
 | |
8OPU
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Sulfamethoxazole (Fragment-B-E1) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystallographic fragment-binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate-channeling path between them.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7PDR
 
 | | Crystal structure of Lymnaea stagnalis Acetylcholine-binding protein (Ls-AChBP) Q55R/M114V double mutant complexed with Dichloromezotiaz | | Descriptor: | 3-[3,5-bis(chloranyl)phenyl]-1-[(2-chloranyl-1,3-thiazol-5-yl)methyl]-9-methyl-pyrido[1,2-a]pyrimidine-2,4-dione, Acetylcholine-binding protein | | Authors: | Montgomery, M.G. | | Deposit date: | 2021-08-06 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural Biology-Guided Design, Synthesis, and Biological Evaluation of Novel Insect Nicotinic Acetylcholine Receptor Orthosteric Modulators.
J.Med.Chem., 65, 2022
|
|
7PE5
 
 | |
8OTL
 
 | | structure of InhA from Mycobacterium tuberculosis in complex with 5-(((4-(2-hydroxyphenoxy)benzyl)(octyl)amino)methyl)-2-phenoxyphenol | | Descriptor: | 1,2-ETHANEDIOL, 5-[[octyl-[[4-(2-oxidanylphenoxy)phenyl]methyl]amino]methyl]-2-phenoxy-phenol, ACETATE ION, ... | | Authors: | Tamhaev, R, Maveyraud, L, Chebaiki, M, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Exploring the plasticity of the InhA substrate-binding site using new diaryl ether inhibitors.
Bioorg.Chem., 143, 2023
|
|
