4ZO8
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorose | | Descriptor: | Lin1840 protein, MAGNESIUM ION, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4G1H
 
 | |
5XF5
 
 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,S-configuration) | | Descriptor: | (1S,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
1HH8
 
 | | The active N-terminal region of p67phox: Structure at 1.8 Angstrom resolution and biochemical characterizations of the A128V mutant implicated in chronic granulomatous disease | | Descriptor: | CITRATE ANION, NEUTROPHIL CYTOSOL FACTOR 2 | | Authors: | Grizot, S, Fieschi, F, Dagher, M.-C, Pebay-Peyroula, E. | | Deposit date: | 2000-12-21 | | Release date: | 2001-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active N-Terminal Region of P67Phox: Structure at 1.8 Angstrom Resolution and Biochemical Characterizations of the A128V Mutant Implicated in Chronic Granulomatous Disease
J.Biol.Chem., 276, 2001
|
|
1UIY
 
 | | Crystal Structure of Enoyl-CoA Hydratase from Thermus Thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Enoyl-CoA Hydratase, GLYCEROL | | Authors: | Bagautdinov, B, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-24 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2UYV
 
 | |
3UYS
 
 | | Crystal structure of apo human ck1d | | Descriptor: | Casein kinase I isoform delta, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2011-12-06 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor.
J.Med.Chem., 55, 2012
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
2VN7
 
 | | Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea jecorina | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bott, R, Sandgren, M, Hansson, H. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of an Intact Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea Jecorina.
Biochemistry, 47, 2008
|
|
6QDV
 
 | | Human post-catalytic P complex spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Fica, S.M, Oubridge, C, Wilkinson, M.E, Newman, A.J, Nagai, K. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation.
Science, 363, 2019
|
|
4JBG
 
 | | 1.75A resolution structure of a thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
3V3M
 
 | | Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease in Complex with N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide inhibitor. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Jacobs, J, Grum-Tokars, V, Zhou, Y, Turlington, M, Saldanha, S.A, Chase, P, Eggler, A, Dawson, E.S, Baez-Santos, Y.M, Tomar, S, Mielech, A.M, Baker, S.C, Lindsley, C.W, Hodder, P, Mesecar, A, Stauffer, S.R. | | Deposit date: | 2011-12-13 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease.
J.Med.Chem., 56, 2013
|
|
4CAF
 
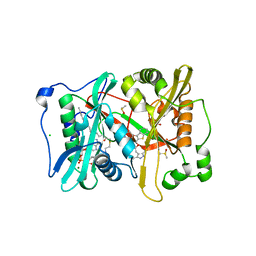 | | Plasmodium vivax N-myristoyltransferase in complex with a benzothiophene inhibitor (compound 34a) | | Descriptor: | 2-oxopentadecyl-CoA, 4-[(2-{5-[(3,5-dimethyl-1H-pyrazol-4-yl)methyl]-1,3,4-oxadiazol-2-yl}-1-benzothiophen-3-yl)oxy]piperidine, AMMONIUM ION, ... | | Authors: | Rackham, M.D, Brannigan, J.A, Rangachari, K, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of High Affinity Inhibitors of Plasmodium Falciparum and Plasmodium Vivax N-Myristoyltransferases Directed by Ligand Efficiency Dependent Lipophilicity (Lelp).
J.Med.Chem., 57, 2014
|
|
4FTC
 
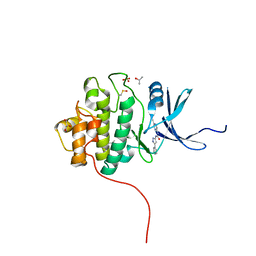 | | Crystal Structure of the CHK1 | | Descriptor: | 1-(5-cyanopyrazin-2-yl)-3-(5-phenyl-1H-pyrazol-3-yl)urea, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
108D
 
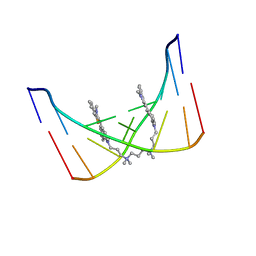 | | THE SOLUTION STRUCTURE OF A DNA COMPLEX WITH THE FLUORESCENT BIS INTERCALATOR TOTO DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | 1,1-(4,4,8,8-TETRAMETHYL-4,8-DIAZAUNDECAMETHYLENE)-BIS-4-3-METHYL-2,3-DIHYDRO-(BENZO-1,3-THIAZOLE)-2-METHYLIDENE)-QUINOLINIUM, DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | Authors: | Spielmann, H.P, Wemmer, D.E, Jacobsen, J.P. | | Deposit date: | 1995-01-31 | | Release date: | 1995-06-03 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy.
Biochemistry, 34, 1995
|
|
2P7N
 
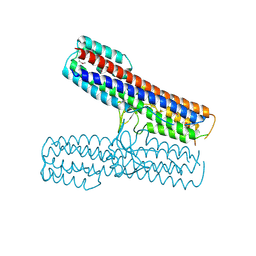 | | Crystal structure of the Pathogenicity island 1 effector protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESGC) target CvR69. | | Descriptor: | Pathogenicity island 1 effector protein | | Authors: | Benach, J, Abashidze, M, Seetharaman, J, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Pathogenicity island 1 effector protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESGC) target CvR69.
To be Published
|
|
7H7N
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z915492990 (CHIKV_MacB-x0892) | | Descriptor: | 1-methyl-N-[(thiophen-2-yl)methyl]-1H-pyrazole-5-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
3F7R
 
 | |
4BVP
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with heptamolybdate and octamolybdate | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2013-06-27 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structures of Legionella Pneumophila Ntpdase1 in Complex with Polyoxometallates.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3F99
 
 | | W354F Yersinia enterocolitica PTPase apo form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase yopH | | Authors: | Brandao, T.A.S, Robinson, H, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2008-11-13 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Impaired acid catalysis by mutation of a protein loop hinge residue in a YopH mutant revealed by crystal structures.
J.Am.Chem.Soc., 131, 2009
|
|
3VHE
 
 | | Crystal structure of human VEGFR2 kinase domain with a novel pyrrolopyrimidine inhibitor. | | Descriptor: | 1-{2-fluoro-4-[(5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy]phenyl}-3-[3-(trifluoromethyl)phenyl]urea, Vascular endothelial growth factor receptor 2 | | Authors: | Oguro, Y, Miyamoto, N, Okada, K, Takagi, T, Iwata, H, Awazu, Y, Miki, H, Hori, A, Kamiyama, K, Imanura, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, synthesis, and evaluation of 5-methyl-4-phenoxy-5H-pyrrolo[3,2-d]pyrimidine derivatives: novel VEGFR2 kinase inhibitors binding to inactive kinase conformation.
Bioorg.Med.Chem., 18, 2010
|
|
4G1J
 
 | | Sortase C1 of GBS Pilus Island 1 | | Descriptor: | Sortase family protein | | Authors: | Cozzi, R, Prigozhin, D.M, Alber, T. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for group B streptococcus pilus 1 sortases C regulation and specificity.
Plos One, 7, 2012
|
|
3VAG
 
 | | Structure of U2AF65 variant with BrU3C2 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*U*CP*(BRU)P*UP*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3EYX
 
 | | Crystal structure of Carbonic Anhydrase Nce103 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Carbonic anhydrase, ... | | Authors: | Teng, Y.B, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2008-10-22 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into the substrate tunnel of Saccharomyces cerevisiae carbonic anhydrase Nce103.
Bmc Struct.Biol., 9, 2009
|
|
4OH7
 
 | |
