1Z3V
 
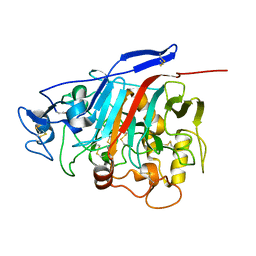 | | Structure of Phanerochaete chrysosporium cellobiohydrolase Cel7D (CBH58) in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, cellulase | | Authors: | Ubhayasekera, W, Munoz, I.G, Stahlberg, J, Mowbray, S.L. | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors
Febs J., 272, 2005
|
|
1Z3W
 
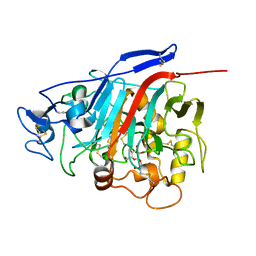 | | Structure of Phanerochaete chrysosporium cellobiohydrolase Cel7D (CBH58) in complex with cellobioimidazole | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, cellulase | | Authors: | Ubhayasekera, W, Vasella, A, Stahlberg, J, Mowbray, S.L. | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors
Febs J., 272, 2005
|
|
1Z3X
 
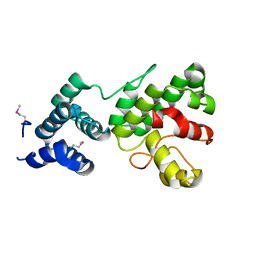 | | Structure of Gun4 from Thermosynechococcus elongatus | | Descriptor: | putative cytidylyltransferase | | Authors: | Davison, P.A, Schubert, H.L, Reid, J.D, Iorg, C.D, Robinson, H, Hill, C.P, Hunter, C.N. | | Deposit date: | 2005-03-14 | | Release date: | 2005-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biochemical Characterization of Gun4 Suggests a Mechanism for Its Role in Chlorophyll Biosynthesis(,).
Biochemistry, 44, 2005
|
|
1Z3Y
 
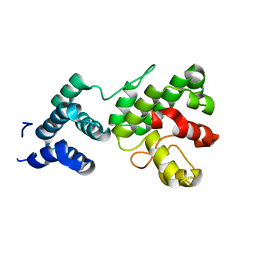 | | Structure of Gun4-1 from Thermosynechococcus elongatus | | Descriptor: | putative cytidylyltransferase | | Authors: | Davison, P.A, Schubert, H.L, Reid, J.D, Iorg, C.D, Robinson, H, Hill, C.P, Hunter, C.N. | | Deposit date: | 2005-03-14 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of Gun4 Suggests a Mechanism for Its Role in Chlorophyll Biosynthesis(,).
Biochemistry, 44, 2005
|
|
1Z3Z
 
 | | The crystal structure of a DGD mutant: Q52A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fogle, E.J, Liu, W, Toney, M.D. | | Deposit date: | 2005-03-14 | | Release date: | 2006-01-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of q52 in catalysis of decarboxylation and transamination in dialkylglycine decarboxylase.
Biochemistry, 44, 2005
|
|
1Z40
 
 | | AMA1 from Plasmodium falciparum | | Descriptor: | CHLORIDE ION, apical membrane antigen 1 precursor | | Authors: | Bai, T, Becker, M, Gupta, A, Strike, P, Murphy, V.J, Anders, R.F, Batchelor, A.H. | | Deposit date: | 2005-03-14 | | Release date: | 2005-08-16 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Z41
 
 | | Crystal structure of oxidized YqjM from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable NADH-dependent flavin oxidoreductase yqjM, SULFATE ION | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z42
 
 | | Crystal structure of oxidized YqjM from Bacillus subtilis complexed with p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE, Probable NADH-dependent flavin oxidoreductase yqjM, ... | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z43
 
 | |
1Z44
 
 | | Crystal structure of oxidized YqjM from Bacillus subtilis complexed with p-nitrophenol | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-NITROPHENOL, Probable NADH-dependent flavin oxidoreductase yqjM, ... | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z45
 
 | | Crystal structure of the gal10 fusion protein galactose mutarotase/UDP-galactose 4-epimerase from Saccharomyces cerevisiae complexed with NAD, UDP-glucose, and galactose | | Descriptor: | GAL10 bifunctional protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2005-03-15 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The molecular architecture of galactose mutarotase/UDP-galactose 4-epimerase from Saccharomyces cerevisiae.
J.Biol.Chem., 280, 2005
|
|
1Z47
 
 | | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius | | Descriptor: | CHLORIDE ION, putative ABC-transporter ATP-binding protein | | Authors: | Scheffel, F, Demmer, U, Warkentin, E, Huelsmann, A, Schneider, E, Ermler, U. | | Deposit date: | 2005-03-15 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius
Febs Lett., 579, 2005
|
|
1Z48
 
 | | Crystal structure of reduced YqjM from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable NADH-dependent flavin oxidoreductase yqjM | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z4A
 
 | |
1Z4E
 
 | |
1Z4H
 
 | | The response regulator TorI belongs to a new family of atypical excisionase | | Descriptor: | Tor inhibition protein | | Authors: | Elantak, L, Ansaldi, M, Guerlesquin, F, Mejean, V, Morelli, X. | | Deposit date: | 2005-03-16 | | Release date: | 2005-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and genetic analyses reveal a key role in prophage excision for the TorI response regulator inhibitor
J.Biol.Chem., 280, 2005
|
|
1Z4I
 
 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with deoxyribouridine 5'-monophosphate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5'(3')-deoxyribonucleotidase, GLYCEROL, ... | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1Z4J
 
 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with uridine 2'-monophosphate | | Descriptor: | 5'(3')-deoxyribonucleotidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1Z4K
 
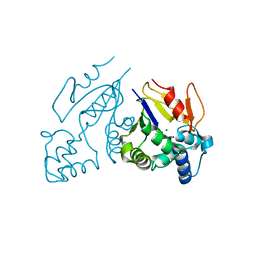 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with thymidine 3'-monophosphate | | Descriptor: | 5'(3')-deoxyribonucleotidase, MAGNESIUM ION, THYMIDINE-3'-PHOSPHATE | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1Z4L
 
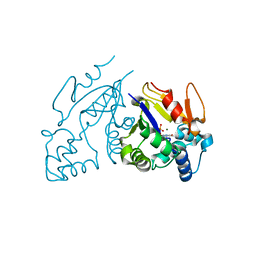 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with thymidine 5'-monophosphate | | Descriptor: | 5'(3')-deoxyribonucleotidase, MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1Z4M
 
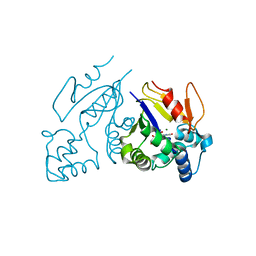 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with uridine 5'-monophosphate | | Descriptor: | 5'(3')-deoxyribonucleotidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1Z4N
 
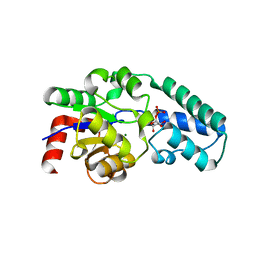 | | Structure of beta-phosphoglucomutase with inhibitor bound alpha-galactose 1-phosphate cocrystallized with Fluoride | | Descriptor: | 1-O-phosphono-alpha-D-galactopyranose, Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Tremblay, L.W, Zhang, G, Dai, J, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-03-16 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Chemical Confirmation of a Pentavalent Phosphorane in Complex with beta-Phosphoglucomutase
J.Am.Chem.Soc., 127, 2005
|
|
1Z4O
 
 | | Structure of beta-phosphoglucomutase with inhibitor bound alpha-galactose 1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-galactopyranose, Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Tremblay, L.W, Zhang, G, Dai, J, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-03-16 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical Confirmation of a Pentavalent Phosphorane in Complex with beta-Phosphoglucomutase
J.Am.Chem.Soc., 127, 2005
|
|
1Z4P
 
 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with deoxyriboguanosine 5'-monophosphate | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, 5'(3')-deoxyribonucleotidase, MAGNESIUM ION | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1Z4Q
 
 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with 2',3'-dideoxy-2',3-didehydrothymidine 5'-monophosphate (d4T-MP) | | Descriptor: | 5'(3')-deoxyribonucleotidase, MAGNESIUM ION, [(5R)-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)-2,5-DIHYDROFURAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
