3DVQ
 
 | |
3DW1
 
 | |
3DVK
 
 | | Crystal Structure of Ca2+/CaM-CaV2.3 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent R-type calcium channel subunit alpha-1E | | Authors: | Kim, E.Y, Rumpf, C.H, Fujiwara, Y, Cooley, E.S, Van Petegem, F, Minor, D.L. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Ca(V)2 Ca(2+)/CaM-IQ Domain Complexes Reveal Binding Modes that Underlie Calcium-Dependent Inactivation and Facilitation.
Structure, 16, 2008
|
|
3DYI
 
 | |
3DZO
 
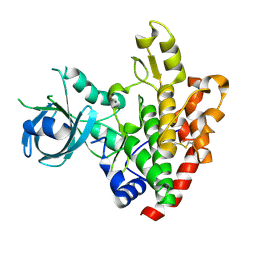 | | Crystal structure of a rhoptry kinase from toxoplasma gondii | | Descriptor: | MAGNESIUM ION, Rhoptry kinase domain | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Ni, S, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sibley, D, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
3DWY
 
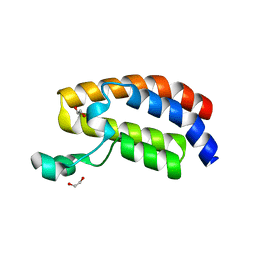 | | Crystal Structure of the Bromodomain of Human CREBBP | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Fedorov, O, Karim, R, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-23 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3E1W
 
 | |
3DQN
 
 | |
3E19
 
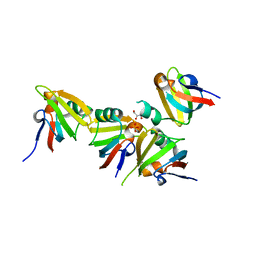 | | Crystal Structure of Iron Uptake Regulatory Protein (FeoA) Solved by Sulfur SAD in a Monoclinic Space Group | | Descriptor: | FeoA, GLYCEROL, PHOSPHATE ION | | Authors: | Hughes, R.C, Li, Y, Wang, B.-C, Liu, Z.-J, Ng, J.D. | | Deposit date: | 2008-08-02 | | Release date: | 2008-12-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Structure Determination of Iron Uptake Regulatory
Protein (FeoA) by Sulfur SAD in a Monoclinic Space Group
To be Published
|
|
3DV6
 
 | | Crystal structure of SAG506-01, tetragonal, crystal 2 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-18 | | Release date: | 2008-12-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3DX7
 
 | | Crystal Structure of HLA-B*4403 presenting 10mer EBV antigen | | Descriptor: | ACETATE ION, Beta-2-microglobulin, EBV decapeptide epitope, ... | | Authors: | Archbold, J.K, Ely, L.K, Rossjohn, J. | | Deposit date: | 2008-07-23 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition.
J.Exp.Med., 206, 2009
|
|
3DXN
 
 | | Crystal structure of the calcium-dependent kinase from toxoplasma gondii, 541.m00134, kinase domain. | | Descriptor: | Calmodulin-like domain protein kinase isoform 3 | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Cossar, D, Wasney, G, Lin, Y.H, Hassani, A, Ali, A, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wikstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-24 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the calcium-dependent kinase from toxoplasma gondii, 541.m00134, kinase domain.
To be Published
|
|
3EK7
 
 | | Calcium-saturated GCaMP2 dimer | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Akerboom, J, Velez Rivera, J.D, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of the GCaMP Calcium Sensor Reveal the Mechanism of Fluorescence Signal Change and Aid Rational Design
J.Biol.Chem., 284, 2009
|
|
3E2H
 
 | |
3E28
 
 | |
3E3Z
 
 | | Crystal structure of bovine coupling Factor B bound with phenylarsine oxide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP synthase subunit s, mitochondrial, ... | | Authors: | Stroud, R.M, Lee, J.K, Belogrudov, G.I. | | Deposit date: | 2008-08-08 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of bovine mitochondrial factor B at 0.96-A resolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3E74
 
 | |
3E83
 
 | | Crystal Structure of the the open NaK channel pore | | Descriptor: | CESIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Jiang, Y, Alam, A. | | Deposit date: | 2008-08-19 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of ion selectivity in the NaK channel
Nat.Struct.Mol.Biol., 16, 2009
|
|
3E8H
 
 | |
3E9C
 
 | |
3EA3
 
 | | Crystal Structure of the Y246S/Y247S/Y248S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, MANGANESE (II) ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Modulation of Bacillus thuringiensis Phosphatidylinositol-specific Phospholipase C Activity by Mutations in the Putative Dimerization Interface.
J.Biol.Chem., 284, 2009
|
|
3EI0
 
 | |
3EKT
 
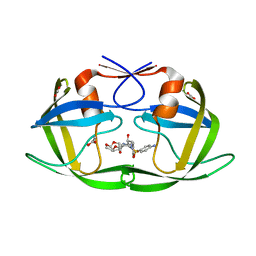 | | Crystal Structure of the inhibitor Darunavir (DRV) in complex with a multi-drug resistant HIV-1 protease variant (L10F/G48V/I54V/V64I/V82A) (Refer: FLAP+ in citation.) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N.M, Bandaranayake, R.M. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3E2L
 
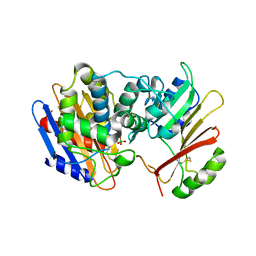 | | Crystal Structure of the KPC-2 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) | | Descriptor: | Beta-lactamase inhibitory protein, Carbapenemase | | Authors: | Hanes, M.S, Jude, K.M, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein
Biochemistry, 48, 2009
|
|
3E55
 
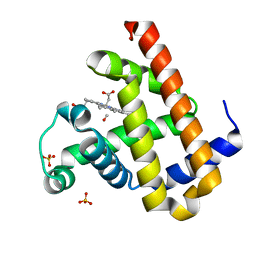 | | Carbonmonoxy Sperm Whale Myoglobin at 100 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-13 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
