6I56
 
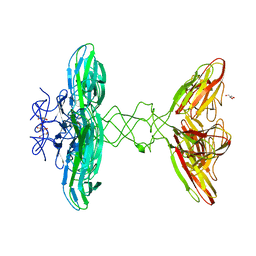 | | Crystal structure of PBSX exported protein XepA | | Descriptor: | GLYCEROL, Phage-like element PBSX protein XepA | | Authors: | Hakansson, M, Svensson, L.A, Welin, M, Al-Karadaghi, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of the Bacillus subtilis prophage lytic cassette proteins XepA and YomS.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
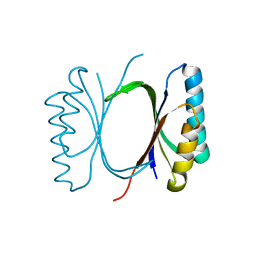
6I69
 
 | |
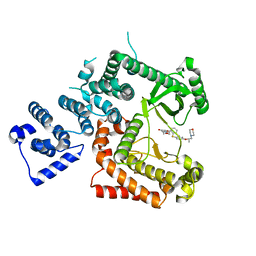
6I3U
 
 | |
6I7I
 
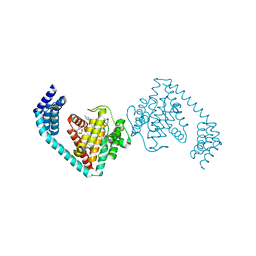 | | Crystal structure of dimeric FICD mutant K256A complexed with MgATP | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6ECA
 
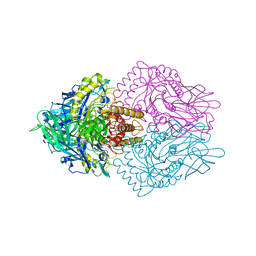 | | Lactobacillus rhamnosus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Pellock, S.J, Bhatt, A.P, Bivins, M.M, Walton, W.G, Tran, B.N.T, Wei, L, Snider, M.C, Cesmat, A.P, Tripathy, A, Erie, D.A, Redinbo, M.R.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
6ENE
 
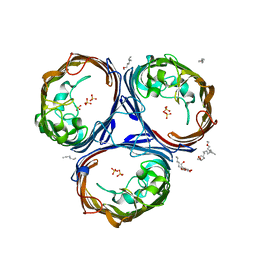 | | OmpF orthologue from Enterobacter cloacae (OmpE35) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein (Porin), SULFATE ION, ... | | Authors: | van den Berg, B, Abellon-Ruiz, J, Basle, A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
6EO3
 
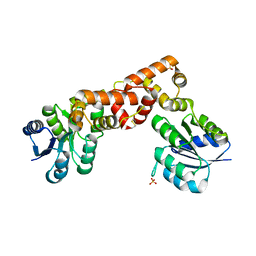 | |
6ERN
 
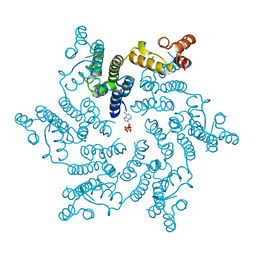 | | HIV Hexamer with ligand | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Gag polyprotein | | Authors: | James, L.C. | | Deposit date: | 2017-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | HIV hexamer
To Be Published
|
|
6KNK
 
 | | Crystal structure of SbnH in complex with citryl-diaminoethane | | Descriptor: | (2S)-2-{2-[(2-AMINOETHYL)AMINO]-2-OXOETHYL}-2-HYDROXYBUTANEDIOIC ACID, (2~{S})-2-[2-[2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]ethylamino]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, PHOSPHATE ION, ... | | Authors: | Tang, J, Ju, Y, Zhou, H. | | Deposit date: | 2019-08-05 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis.
J.Mol.Biol., 431, 2019
|
|
6EO6
 
 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(2-(1H-indol-3-yl)acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA63A - TBA MODIFIED APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
6K6V
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JY1
 
 | |
6ERM
 
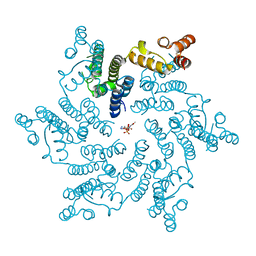 | | HIV Hexamer with ligand | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, Gag polyprotein | | Authors: | James, L.C. | | Deposit date: | 2017-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV hexamer
To Be Published
|
|
6E8C
 
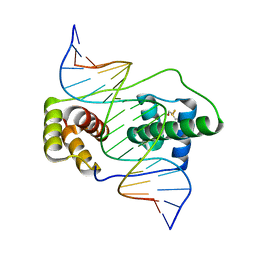 | | Crystal structure of the double homeodomain of DUX4 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), Double homeobox protein 4 | | Authors: | Lee, J.K, Bosnakovski, D, Toso, E.A, Dinh, T, Banerjee, S, Bohl, T.E, Shi, K, Kurahashi, K, Kyba, M, Aihara, H. | | Deposit date: | 2018-07-27 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Cell Rep, 25, 2018
|
|
6E99
 
 | |
6K9S
 
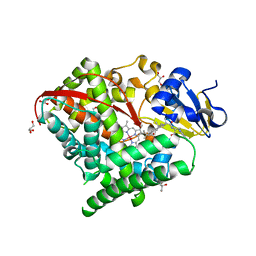 | | Structure of the Carbonylruthenium Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CARBON MONOXIDE, ... | | Authors: | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-06-17 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6EA7
 
 | | Structure of EBOV GPcl in complex with the pan-ebolavirus mAb ADI-15878 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | West, B.R, Moyer, C.L, King, L.B, Fusco, M.L, Milligan, J.C, Hui, S, Saphire, E.O. | | Deposit date: | 2018-08-02 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Structural Basis of Pan-Ebolavirus Neutralization by a Human Antibody against a Conserved, yet Cryptic Epitope.
MBio, 9, 2018
|
|
6K3Q
 
 | | Crystal Structure of P450BM3 with N-(3-cyclohexylpropanoyl)-L-prolyl-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-1-(3-cyclohexylpropanoyl)pyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Shoji, O, Yonemura, K. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Systematic Evolution of Decoy Molecules for the Highly Efficient Hydroxylation of Benzene and Small Alkanes Catalyzed by Wild-Type Cytochrome P450BM3
Acs Catalysis, 10, 2020
|
|
6EAH
 
 | |
6EBC
 
 | | OhrB (Organic Hydroperoxide Resistance protein) wild type from Chromobacterium violaceum and reduced by DTT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Organic hydroperoxide resistance protein | | Authors: | Domingos, R.M, Teixeira, R.D, Alegria, T.G.P, Vieira, P.S, Murakami, M.T, Netto, L.E.S. | | Deposit date: | 2018-08-06 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Substrate and product-assisted catalysis: molecular aspects behind structural switches along Organic Hydroperoxide Resistance Protein catalytic cycle
Acs Catalysis, 2020
|
|
6KEQ
 
 | | Crystal structure of D113A mutant of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in apo-form | | Descriptor: | Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6ED1
 
 | | Bacteroides dorei Beta-glucuronidase | | Descriptor: | Glycosyl hydrolase family 2, sugar binding domain protein, SODIUM ION | | Authors: | Biernat, K.A, Redinbo, M.R. | | Deposit date: | 2018-08-08 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
6EEU
 
 | | Structure of class II HMG-CoA reductase from Delftia acidovorans | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-3-methylglutaryl coenzyme A reductase, SULFATE ION | | Authors: | Ragwan, E.R, Arai, E, Kung, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | New Crystallographic Snapshots of Large Domain Movements in Bacterial 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase.
Biochemistry, 57, 2018
|
|
6KMK
 
 | | Crystal structure of hydrogen peroxide bound bovine lactoperoxidase at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Singh, P.K, Sirohi, H.V, Bhusan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of hydrogen peroxide bound bovine lactoperoxidase at 2.3 A resolution
To Be Published
|
|
6EGY
 
 | | Crystal structure of cytochrome c in complex with mono-PEGylated sulfonatocalix[4]arene | | Descriptor: | Cytochrome c iso-1, HEME C, SULFATE ION, ... | | Authors: | Mummidivarapu, V.V.S, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent PEGylation via Sulfonatocalix[4]arene-A Crystallographic Proof.
Bioconjug.Chem., 29, 2018
|
|
