6NUS
 
 | |
5I08
 
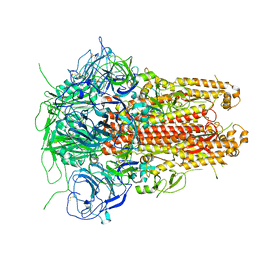 | | Prefusion structure of a human coronavirus spike protein | | Descriptor: | Spike glycoprotein,Foldon chimera | | Authors: | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
6GUQ
 
 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with glucose | | Descriptor: | Putative sugar binding protein, beta-D-glucopyranose | | Authors: | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2018-06-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | The crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with glucose
To Be Published
|
|
6GT9
 
 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with galactose | | Descriptor: | Putative sugar binding protein, SULFATE ION, beta-D-galactopyranose | | Authors: | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2018-06-16 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | The crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with galactose
To Be Published
|
|
6GQ0
 
 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus | | Descriptor: | Putative sugar binding protein | | Authors: | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of GanP, a glucose-galactose binding protein from Gebacillus Stearothermophilus
To Be Published
|
|
2DIX
 
 | | Solution structure of the DSRM domain of Protein activator of the interferon-induced protein kinase | | Descriptor: | Interferon-inducible double stranded RNA-dependent protein kinase activator A | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DSRM domain of Protein activator of the interferon-induced protein kinase
To be published
|
|
2YU0
 
 | | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205 | | Descriptor: | Interferon-activable protein 205 | | Authors: | Sato, M, Tochio, N, Koshiba, S, Watanabe, M, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205
To be Published
|
|
6F5M
 
 | | Crystal structure of highly glycosylated human leukocyte elastase in complex with a thiazolidinedione inhibitor | | Descriptor: | 5-[[4-[[(2~{S})-4-methyl-1-oxidanylidene-1-[(2-propylphenyl)amino]pentan-2-yl]carbamoyl]phenyl]methyl]-2-oxidanylidene-1,3-thiazol-1-ium-4-olate, ACETATE ION, Neutrophil elastase, ... | | Authors: | Hochscherf, J, Pietsch, M, Tieu, W, Kuan, K, Hautmann, S, Abell, A, Guetschow, M, Niefind, K. | | Deposit date: | 2017-12-01 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of highly glycosylated human leukocyte elastase in complex with an S2' site binding inhibitor.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5B61
 
 | | Extra-superfolder GFP | | Descriptor: | Green fluorescent protein | | Authors: | Park, H.H, Jang, T.-H, Choi, J.Y. | | Deposit date: | 2016-05-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | The mechanism of folding robustness revealed by the crystal structure of extra-superfolder GFP.
FEBS Lett., 591, 2017
|
|
2DLL
 
 | | Solution structure of the IRF domain of human interferon regulator factors 4 | | Descriptor: | Interferon regulatory factor 4 | | Authors: | Zhang, H.P, Kurosaki, C, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF domain of human interferon regulator factors 4
To be published
|
|
4N6X
 
 | | Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1/Chemokine Receptor CXCR2 fusion protein | | Authors: | Lu, G, Wu, Y, Jiang, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.051 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
5A8L
 
 | | Human eRF1 and the hCMV nascent peptide in the translation termination complex | | Descriptor: | 28S RIBOSOMAL RNA, 60S RIBOSOMAL PROTEIN L12, 60S RIBOSOMAL PROTEIN L17, ... | | Authors: | Matheisl, S, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2015-07-16 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Human Translation Termination Complex.
Nucleic Acids Res., 43, 2015
|
|
4MPA
 
 | | Crystal structure of NHERF1-CXCR2 signaling complex in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1, ... | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
2KTV
 
 | |
1X48
 
 | | Solution structure of the second DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | He, F, Muto, Y, Inoue, M, Tarada, T, Shirouzu, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase
To be Published
|
|
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-07-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
2HST
 
 | |
2OZF
 
 | | The crystal structure of the 2nd PDZ domain of the human NHERF-1 (SLC9A3R1) | | Descriptor: | Ezrin-radixin-moesin-binding phosphoprotein 50 | | Authors: | Phillips, C, Papagrigoriou, E, Gileadi, C, Fedorov, O, Elkins, J, Berridge, G, Turnbull, A.P, Gileadi, O, Schoch, G, Smee, C, Bray, J, Savitsky, P, Uppenberg, J, von Delft, F, Gorrec, F, Umeano, C, Salah, E, Colebrook, S, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 2nd PDZ domain of the human NHERF-1 (SLC9A3R1)
To be Published
|
|
1X49
 
 | | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase
To be Published
|
|
2MQ6
 
 | |
2MQ9
 
 | |
2HJ8
 
 | | Solution NMR structure of the C-terminal domain of the interferon alpha-inducible ISG15 protein from Homo sapiens. Northeast Structural Genomics target HR2873B | | Descriptor: | Interferon-induced 17 kDa protein | | Authors: | Aramini, J.M, Ho, C.K, Yin, C, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal domain of the interferon alpha-inducible ISG15 protein from Homo sapiens. Northeast Structural Genomics target HR2873B.
To be Published
|
|
3HIP
 
 | |
3QG7
 
 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Antibody Heavy Chain, AP4-24H11 Antibody Light Chain, HEXAETHYLENE GLYCOL, ... | | Authors: | Kirchdoerfer, R.K, Kaufmann, G.F, Janda, J.D, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
1KIB
 
 | | cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in the form of an oblate shell | | Descriptor: | HEME C, cytochrome c6 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Krogmann, D, Yeates, T.O. | | Deposit date: | 2001-12-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in a nearly symmetric shell.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
