6DYG
 
 | | Fe(II)-bound structure of the engineered cyt cb562 variant, CH3Y | | Descriptor: | FE (III) ION, HEME C, MAGNESIUM ION, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
7MP4
 
 | | Crystal structure of Epiphyas postvittana antennal carboxylesterase 24 | | Descriptor: | Carboxylesterase-24, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hamiaux, C, Carraher, C. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of an antennally-expressed carboxylesterase suggests lepidopteran odorant degrading enzymes are broadly tuned
Curr Res Insect Sci, 3, 2023
|
|
8UPZ
 
 | |
6H9P
 
 | | MamM CTD E289D - Manganese form | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, Magnetosome protein MamM, ... | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-08-05 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MamM CTD E289D - Manganese form
To Be Published
|
|
8U2V
 
 | |
7MNJ
 
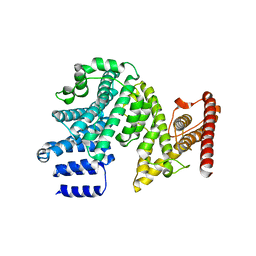 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 145-673) | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Patke, A, Dasso, M, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
6DZX
 
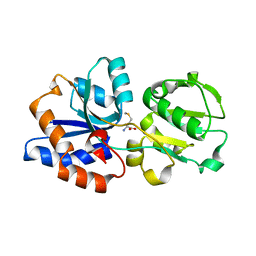 | | Crystal structure of the N. meningitides methionine-binding protein in its D-methionine bound conformation. | | Descriptor: | D-METHIONINE, Lipoprotein | | Authors: | Nguyen, P.T, Lai, J.Y, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers.
Protein Sci., 28, 2019
|
|
6H9Z
 
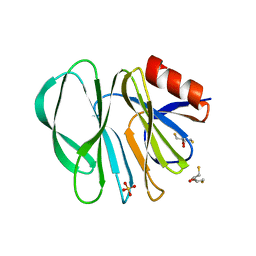 | | Molecular bases of histo-blood group antigen recognition by the most common human rotavirus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Outer capsid protein VP4, SULFATE ION | | Authors: | Ciges-Tomas, J.R, Gozalbo-Rovira, R, Vila-Vicent, S, Buesa, J, Santiso-Bellon, C, Monedero, V, Yebra, M.J, Rodriguez-Diaz, J, Marina, A. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens.
Plos Pathog., 15, 2019
|
|
8U91
 
 | |
6E0N
 
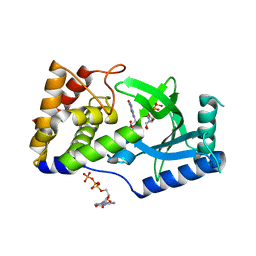 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase with GTP and Apcpp | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
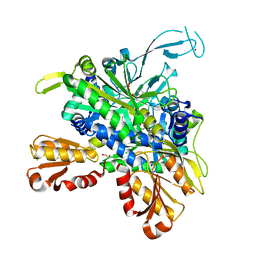
8U2Q
 
 | |
6HYE
 
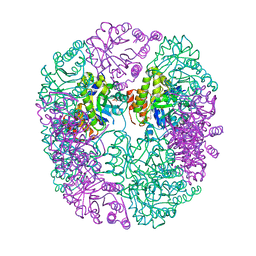 | | PDX1.2/PDX1.3 complex (PDX1.3:K97A) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8U2T
 
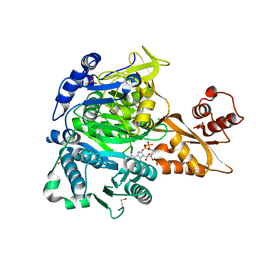 | |
6HAO
 
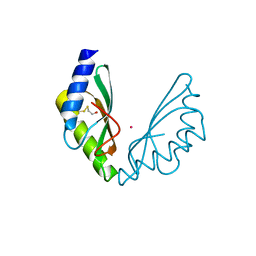 | | MamM CTD H264E - Cadmium form 2 | | Descriptor: | BETA-MERCAPTOETHANOL, CADMIUM ION, Magnetosome protein MamM, ... | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MamM CTD H264E - Cadmium form 2
To Be Published
|
|
8U92
 
 | |
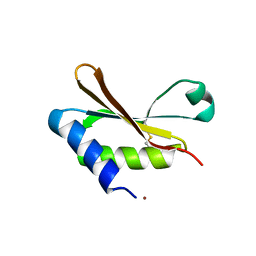
6HAS
 
 | |
6E2A
 
 | | Crystal structure of NADH:quinone reductase PA1024 from Pseudomonas aeruginosa PAO1 in complex with NAD+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Reis, R.A.G, Ball, J, Agniswamy, J, Weber, I, Gadda, G. | | Deposit date: | 2018-07-10 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Steric hindrance controls pyridine nucleotide specificity of a flavin-dependent NADH:quinone oxidoreductase.
Protein Sci., 28, 2019
|
|
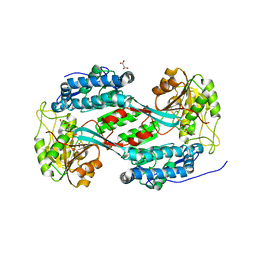
8U9B
 
 | |
8U2U
 
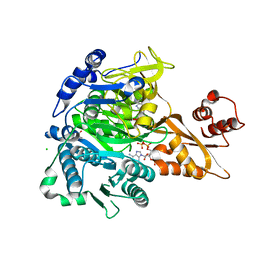 | |
8U2R
 
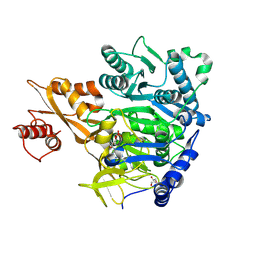 | |
8U98
 
 | |
6HBT
 
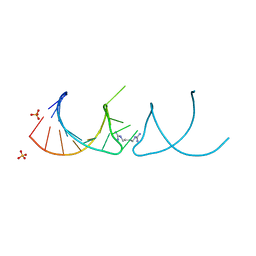 | |
8U90
 
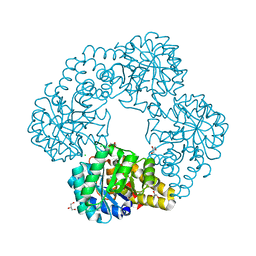 | |
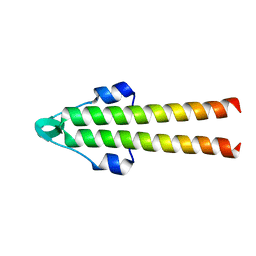
7O39
 
 | |
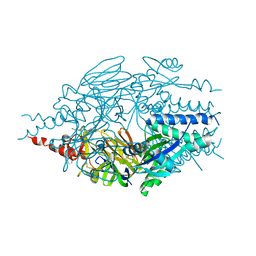
6EAG
 
 | |
