4NR2
 
 | | Crystal structure of STK4 (MST1) SARAH domain | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase 4 | | Authors: | Chaikuad, A, Krojer, T, Kopec, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of STK4 (MST1) SARAH domain
To be Published
|
|
3JWA
 
 | | Crystal structure of L-methionine gamma-lyase from Citrobacter freundii with methionine phosphinate | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHINIC ACID, DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase | | Authors: | Revtovish, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Three-dimensional structures of noncovalent complexes of Citrobacter freundii methionine gamma-lyase with substrates.
Biochemistry Mosc., 76, 2011
|
|
5UKY
 
 | | DHp domain of PhoR of M. tuberculosis - native data | | Descriptor: | 1,2-ETHANEDIOL, ATP-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, S. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Asymmetric Structure of the Dimerization Domain of PhoR, a Sensor Kinase Important for the Virulence of Mycobacterium tuberculosis.
ACS Omega, 2, 2017
|
|
7TN5
 
 | |
5AOB
 
 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-butyrate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
6ROD
 
 | |
7TN6
 
 | |
2QDY
 
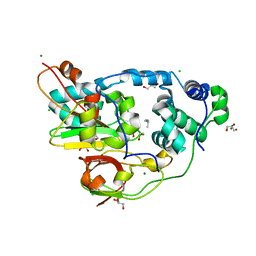 | | Crystal Structure of Fe-type NHase from Rhodococcus erythropolis AJ270 | | Descriptor: | 2-METHYLPROPAN-1-AMINE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Song, L, Shi, J, Xue, Z, Wang, M.-X, Qian, S. | | Deposit date: | 2007-06-22 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution X-ray molecular structure of the nitrile hydratase from Rhodococcus erythropolis AJ270 reveals posttranslational oxidation of two cysteines into sulfinic acids and a novel biocatalytic nitrile hydration mechanism
Biochem.Biophys.Res.Commun., 362, 2007
|
|
6FND
 
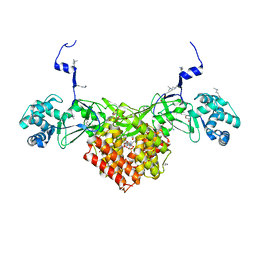 | | Crystal structure of Toxoplasma gondii AKMT | | Descriptor: | 1,2-ETHANEDIOL, Apical complex lysine methyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pivovarova, Y, Dong, G. | | Deposit date: | 2018-02-02 | | Release date: | 2018-11-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of a Novel Dimeric SET Domain Methyltransferase that Regulates Cell Motility.
J. Mol. Biol., 430, 2018
|
|
6RPC
 
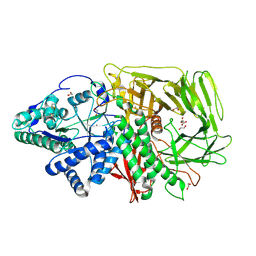 | | GOLGI ALPHA-MANNOSIDASE II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, SUCCINIC ACID, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, Debets, M, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6FPZ
 
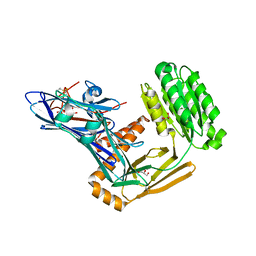 | | Inter-alpha-inhibitor heavy chain 1, D298A | | Descriptor: | ACETATE ION, GLYCEROL, Inter-alpha-trypsin inhibitor heavy chain H1 | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inter-alpha-inhibitor heavy chain-1 has an integrin-like 3D structure mediating immune regulatory activities and matrix stabilization during ovulation
J.Biol.Chem., 2020
|
|
7TN3
 
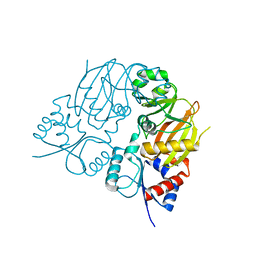 | |
7TN7
 
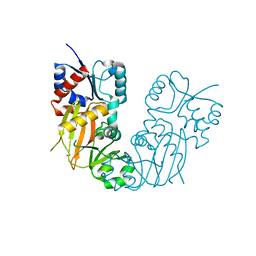 | |
7H1U
 
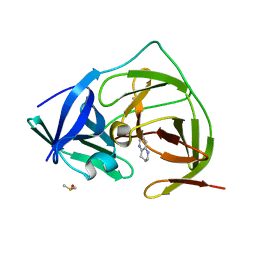 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z212053854 | | Descriptor: | 1,2,3,4-tetrahydro-1,5-naphthyridine, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
6RX4
 
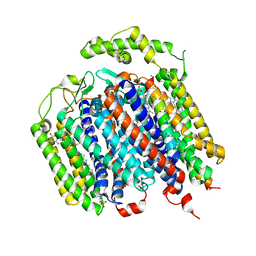 | | THE STRUCTURE OF BD OXIDASE FROM ESCHERICHIA COLI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-I ubiquinol oxidase subunit 1, ... | | Authors: | Rasmussen, T, Boettcher, B, Thesseling, A, Friedrich, T. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Homologous bd oxidases share the same architecture but differ in mechanism.
Nat Commun, 10, 2019
|
|
7THF
 
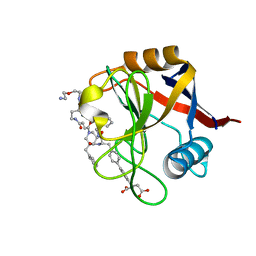 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B53 | | Descriptor: | 3-(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)pentanedioic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
1AMT
 
 | |
5ETV
 
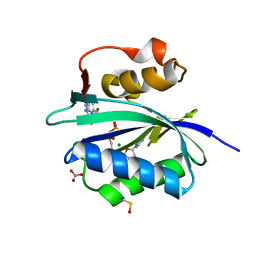 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.72 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[2-(4-bromophenyl)-2-oxidanylidene-ethyl]sulfanyl-1,9-dihydropurin-6-one, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETH
 
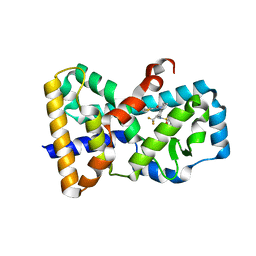 | | RORy in complex with inverse agonist 3. | | Descriptor: | 1-methyl-~{N}-(1-thiophen-2-ylcarbonyl-3,4-dihydro-2~{H}-quinolin-6-yl)-~{N}-[2,2,2-tris(fluoranyl)ethyl]indole-4-sulfonamide, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Discovery of biaryls as ROR gamma inverse agonists by using structure-based design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5NDE
 
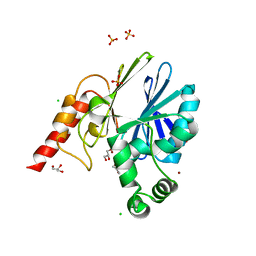 | |
7THD
 
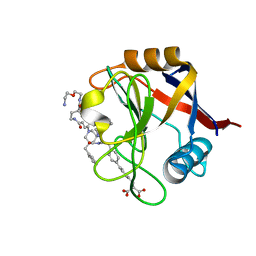 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B52 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, [(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)methyl]propanedioic acid | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
6V0T
 
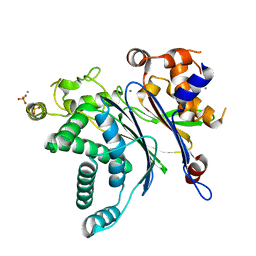 | | Crystal Structure of Catalytic Subunit of Bovine Pyruvate Dehydrogenase Phosphatase 1 - Catalytic Domain | | Descriptor: | MANGANESE (II) ION, SULFATE ION, [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, ... | | Authors: | Guo, Y, Qiu, W, Ernst, S.R, Carroll, D.W, Hackert, M.L. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic subunit of bovine pyruvate dehydrogenase phosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7THC
 
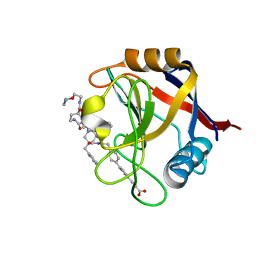 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B25 | | Descriptor: | 3-(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)prop-2-ynoic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
5CBS
 
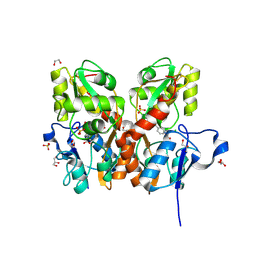 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid at 1.8A resolution | | Descriptor: | (R)-2-amino-3-(3'-hydroxybiphenyl-3-yl)propanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Studies on Aryl-Substituted Phenylalanines: Synthesis, Activity, and Different Binding Modes at AMPA Receptors.
J.Med.Chem., 59, 2016
|
|
9ENQ
 
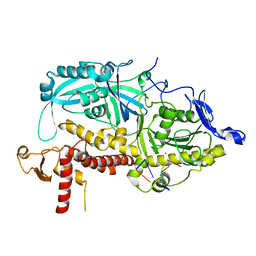 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state active site with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
