1IG3
 
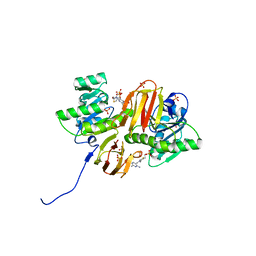 | | Mouse Thiamin Pyrophosphokinase Complexed with Thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, SULFATE ION, thiamin pyrophosphokinase | | Authors: | Timm, D.E, Liu, J, Baker, L.-J, Harris, R.A. | | Deposit date: | 2001-04-16 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of thiamin pyrophosphokinase.
J.Mol.Biol., 310, 2001
|
|
1B5J
 
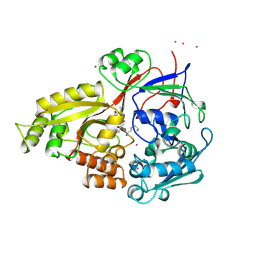 | |
1B04
 
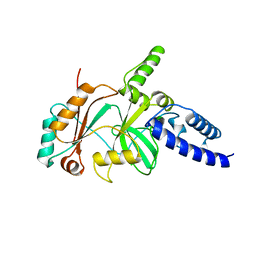 | |
1B2U
 
 | |
1TL1
 
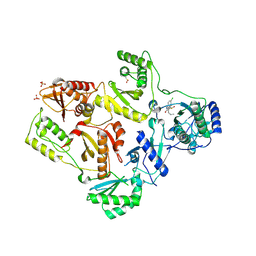 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW451211 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLSULFINYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1G8Z
 
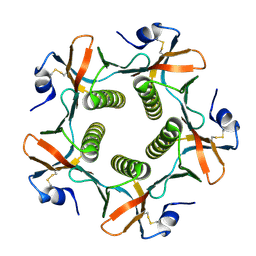 | | HIS57ALA MUTANT OF CHOLERA TOXIN B-PENATMER | | Descriptor: | CHOLERA TOXIN B PROTEIN, beta-D-galactopyranose | | Authors: | Aman, A.T, Fraser, S, Merritt, E.A, Rodigherio, C, Kenny, M. | | Deposit date: | 2000-11-21 | | Release date: | 2001-07-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A mutant cholera toxin B subunit that binds GM1- ganglioside but lacks immunomodulatory or toxic activity.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GC0
 
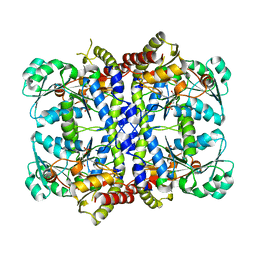 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1GGL
 
 | | HUMAN CELLULAR RETINOL BINDING PROTEIN III | | Descriptor: | PROTEIN (CELLULAR RETINOL-BINDING PROTEIN III) | | Authors: | Calderone, V, Zanotti, G, Folli, C, Ottonello, S, Bolchi, A, Stoppini, M, Berni, R. | | Deposit date: | 2000-08-23 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification, retinoid binding, and x-ray analysis of a human retinol-binding protein.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1AUQ
 
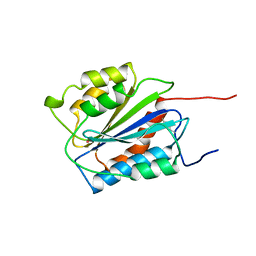 | | A1 DOMAIN OF VON WILLEBRAND FACTOR | | Descriptor: | A1 DOMAIN OF VON WILLEBRAND FACTOR, CADMIUM ION | | Authors: | Emsley, J, Cruz, M, Handin, R, Liddington, R. | | Deposit date: | 1997-09-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the von Willebrand Factor A1 domain and implications for the binding of platelet glycoprotein Ib.
J.Biol.Chem., 273, 1998
|
|
1B51
 
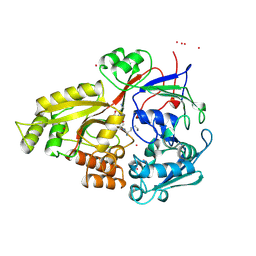 | |
1B9J
 
 | |
1FYA
 
 | |
1UEI
 
 | | Crystal structure of human uridine-cytidine kinase 2 complexed with a feedback-inhibitor, UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, Uridine-cytidine kinase 2 | | Authors: | Suzuki, N.N, Koizumi, K, Fukushima, M, Matsuda, A, Inagaki, F. | | Deposit date: | 2003-05-16 | | Release date: | 2004-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase
STRUCTURE, 12, 2004
|
|
1DZI
 
 | | integrin alpha2 I domain / collagen complex | | Descriptor: | COBALT (II) ION, COLLAGEN, INTEGRIN | | Authors: | Emsley, j, Knight, G, Farndale, R, Barnes, M, Liddington, R. | | Deposit date: | 2000-03-01 | | Release date: | 2001-03-01 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Collagen Recognition by Integrin Alpha2Beta1
Cell(Cambridge,Mass.), 101, 2000
|
|
1G7G
 
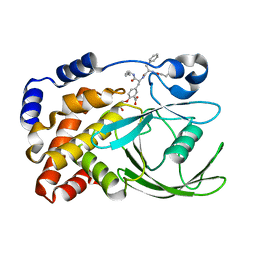 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXES WITH PNU179326 | | Descriptor: | 2-(CARBOXYMETHOXY)-5-[(2S)-2-({(2S)-2-[(3-CARBOXYPROPANOYL)AMINO] -3-PHENYLPROPANOYL}AMINO)-3-OXO-3-(PENTYLAMINO)PROPYL]BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Bleasdale, J.E, Ogg, D, Larsen, S.D. | | Deposit date: | 2000-11-10 | | Release date: | 2001-06-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action.
Biochemistry, 40, 2001
|
|
1DZJ
 
 | | Porcine Odorant Binding Protein Complexed with 2-amino-4-butyl-5-propylselenazole | | Descriptor: | 4-butyl-5-propyl-1,3-selenazol-2-amine, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-03-01 | | Release date: | 2000-12-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexes of Porcine Odorant Binding Protein with Odorant Molecules Belonging to Different Chemical Classes
J.Mol.Biol., 300, 2000
|
|
1G8E
 
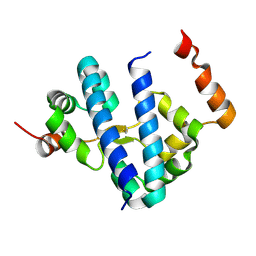 | | CRYSTAL STRUCTURE OF FLHD FROM ESCHERICHIA COLI | | Descriptor: | FLAGELLAR TRANSCRIPTIONAL ACTIVATOR FLHD | | Authors: | Campos, A, Zhang, R.G, Alkire, R.W, Matsumura, P, Westbrook, E.M. | | Deposit date: | 2000-11-17 | | Release date: | 2001-03-07 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the global regulator FlhD from Escherichia coli at 1.8 A resolution.
Mol.Microbiol., 39, 2001
|
|
1E0W
 
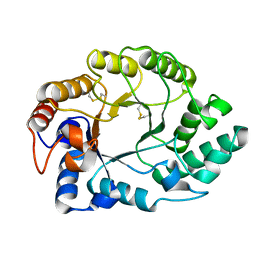 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1TW4
 
 | | Crystal Structure of Chicken Liver Basic Fatty Acid Binding Protein (Bile Acid Binding Protein) Complexed With Cholic Acid | | Descriptor: | CHOLIC ACID, Fatty acid-binding protein | | Authors: | Nichesola, D, Perduca, M, Capaldi, S, Carrizo, M.E, Righetti, P.G, Monaco, H.L. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of chicken liver basic Fatty Acid-binding protein complexed with cholic acid
Biochemistry, 43, 2004
|
|
1XIB
 
 | |
1M4W
 
 | | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa | | Descriptor: | ACETATE ION, GLYCEROL, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hakulinen, N, Turunen, O, Janis, J, Leisola, M, Rouvinen, J. | | Deposit date: | 2002-07-05 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability.
Eur.J.Biochem., 270, 2003
|
|
1DOS
 
 | | STRUCTURE OF FRUCTOSE-BISPHOSPHATE ALDOLASE | | Descriptor: | ALDOLASE CLASS II, AMMONIUM ION, ZINC ION | | Authors: | Blom, N, Tetreault, S, Coulombe, R, Sygusch, J. | | Deposit date: | 1996-06-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel active site in Escherichia coli fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 3, 1996
|
|
1FXY
 
 | | COAGULATION FACTOR XA-TRYPSIN CHIMERA INHIBITED WITH D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | COAGULATION FACTOR XA-TRYPSIN CHIMERA, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide | | Authors: | Hopfner, K.P, Kopetzki, E, Kresse, G.-B, Huber, R, Bode, W, Engh, R.A. | | Deposit date: | 1998-04-22 | | Release date: | 1998-06-17 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | New enzyme lineages by subdomain shuffling.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1MC7
 
 | | Solution Structure of mDvl1 PDZ domain | | Descriptor: | Segment polarity protein dishevelled homolog DVL-1 | | Authors: | Wong, H.-C, Bourdelas, A, Shao, Y, Wu, D, Shi, D.L, Zheng, J. | | Deposit date: | 2002-08-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled
Mol.Cell, 12, 2003
|
|
1DVO
 
 | | THE X-RAY CRYSTAL STRUCTURE OF FINO, A REPRESSOR OF BACTERIAL CONJUGATION | | Descriptor: | FERTILITY INHIBITION PROTEIN O | | Authors: | Ghetu, A.F, Gubbins, M.J, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2000-01-21 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the bacterial conjugation repressor finO.
Nat.Struct.Biol., 7, 2000
|
|
