7NSZ
 
 | | Drosophila PGRP-LB Y78F mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Isoform A of Peptidoglycan-recognition protein LB, SODIUM ION, ... | | Authors: | Orlans, J, Aller, P, Da Silva, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
4DHY
 
 | | Crystal structure of human glucokinase in complex with glucose and activator | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
4MMK
 
 | | Q8A Hfq from Pseudomonas aeruginosa | | Descriptor: | POTASSIUM ION, Protein hfq, SODIUM ION, ... | | Authors: | Murina, V.N, Filimonov, V.V, Melnik, B.S, Uhlein, M, Mueller, U, Weiss, M, Nikulin, A.D. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Effect of conserved intersubunit amino Acid substitutions on hfq protein structure and stability.
Biochemistry Mosc., 79, 2014
|
|
5TX9
 
 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-16 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
4MWJ
 
 | | Anhui N9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWW
 
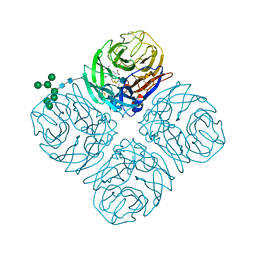 | | Shanghai N9-oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
3KBC
 
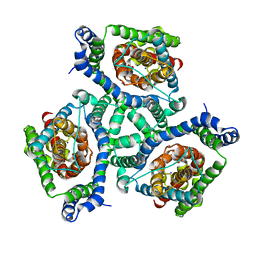 | | Crystal structure of GltPh K55C-A364C mutant crosslinked with divalent mercury | | Descriptor: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, MERCURY (II) ION, ... | | Authors: | Reyes, N, Ginter, C, Boudker, O. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Transport mechanism of a bacterial homologue of glutamate transporters.
Nature, 462, 2009
|
|
7OBZ
 
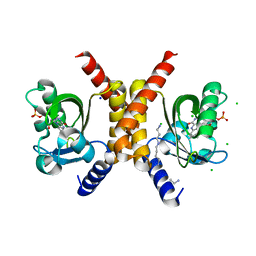 | | Structure of RsLOV D109G | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, LOV protein, ... | | Authors: | Moeglich, A, Krafft, T.G.A, Weyand, M. | | Deposit date: | 2021-04-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Light-Oxygen-Voltage Receptor Integrates Light and Temperature.
J.Mol.Biol., 433, 2021
|
|
7OB0
 
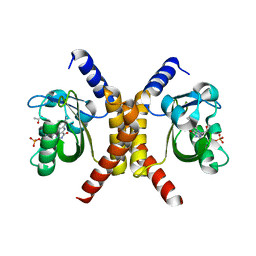 | | Structure of RsLOV d2 variant | | Descriptor: | ACETATE ION, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Moeglich, A, Krafft, T.G.A, Weyand, M. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Light-Oxygen-Voltage Receptor Integrates Light and Temperature.
J.Mol.Biol., 433, 2021
|
|
7NYJ
 
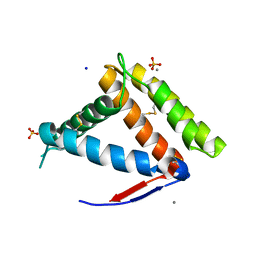 | | Structure of OBP1 from Varroa destructor, form P3<2>21 | | Descriptor: | CALCIUM ION, Odorant Binding Protein 1 from Varoa destructor, form P3<2>21, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-22 | | Release date: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
7NZA
 
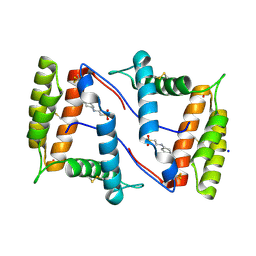 | | Structure of OBP1 from Varroa destructor, form P2<1> | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Odorant Binding Protein from Varroa destructor, form P2<1>, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
7OIL
 
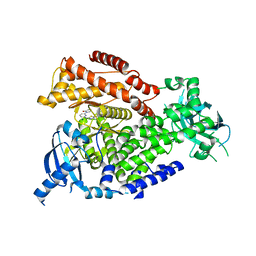 | | mPI3Kd in complex with compound 58 | | Descriptor: | 2-[(1S)-1-cyclopropylethyl]-5-[4-methyl-2-[[6-(2-oxidanylidenepyrrolidin-1-yl)pyridin-2-yl]amino]-1,3-thiazol-5-yl]-7-methylsulfonyl-3H-isoindol-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7O9J
 
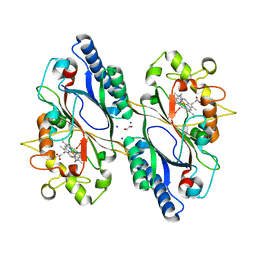 | | Crystal structure of DyP-type peroxidase from Dictyostelium discoideum in complex with an activated form of oxygen | | Descriptor: | 1,2-ETHANEDIOL, DyPA, OXYGEN MOLECULE, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
7OIS
 
 | | mPI3Kd in complex with compound 7 | | Descriptor: | N-[5-[2-[(1S)-1-cyclopropylethyl]-7-[(3-methylsulfonylphenyl)sulfamoyl]-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OIJ
 
 | | mPI3Kd in complex with an inhibitor | | Descriptor: | 6-[[5-[2-[(1S)-1-cyclopropylethyl]-7-(methylsulfamoyl)-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]amino]-N-[3-(dimethylamino)propyl]pyridine-2-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION, ... | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
5TR9
 
 | |
4NYY
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 2 21 21 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6TOP
 
 | | Structure of the PorE C-terminal domain, a protein of T9SS from Porphyromonas gingivalis | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, OmpA family protein, SODIUM ION | | Authors: | Trinh, T.N, Cambillau, C, Roussel, A, Leone, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Type IX secretion system PorE C-terminal domain from Porphyromonas gingivalis in complex with a peptidoglycan fragment.
Sci Rep, 10, 2020
|
|
4DJ4
 
 | | X-ray structure of mutant N211D of bifunctional nuclease TBN1 from Solanum lycopersicum (Tomato) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Nuclease, ... | | Authors: | Koval, T, Stepankova, A, Lipovova, P, Podzimek, T, Matousek, J, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2012-02-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: structural study and reaction-mechanism analysis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6GBN
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Cytophaga hutchinsonii in complex with adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Czyrko, J, Jaskolski, M, Brzezinski, K. | | Deposit date: | 2018-04-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Cytophaga hutchinsonii, a case of combination of crystallographic and non-crystallographic symmetry.
Croatica Chemica Acta, 91, 2018
|
|
5UIM
 
 | | X-ray structure of the FdtF N-formyltransferase from salmonella enteric O60 in complex with folinic acid and TDP-Qui3N | | Descriptor: | Formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, POTASSIUM ION, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
6TRW
 
 | |
6GD1
 
 | | Structure of HuR RRM3 | | Descriptor: | SODIUM ION, Thioredoxin 1,ELAV-like protein 1 | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
5YL8
 
 | | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution
To Be Published
|
|
5UR0
 
 | | Crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Machado, A.T.P, Silva, M, Iulek, J. | | Deposit date: | 2017-02-09 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural studies of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi, the first one from phylum Percolozoa.
Biochim. Biophys. Acta, 1866, 2018
|
|
