3EJN
 
 | |
3EJV
 
 | |
3EN8
 
 | |
3EOF
 
 | |
3ELE
 
 | |
3F9S
 
 | |
3FLJ
 
 | |
3F8X
 
 | |
3FGV
 
 | |
3EZ0
 
 | |
3F7C
 
 | |
3FCN
 
 | |
3FJV
 
 | |
3CVJ
 
 | |
3D7Q
 
 | |
3D1C
 
 | |
3D4E
 
 | |
3A0K
 
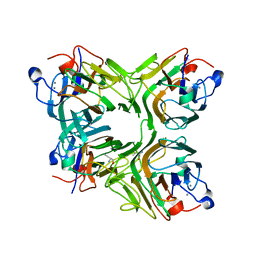 | | Crystal structure of an antiflamatory legume lectin from Cymbosema roseum seeds | | Descriptor: | ALPHA-AMINOBUTYRIC ACID, CALCIUM ION, Cymbosema roseum mannose-specific lectin, ... | | Authors: | Rocha, B.A.M, Delatorre, P, Marinho, E.S, Benevides, R.G, Moura, T.R, Souza, L.A.G, Nascimento, K.S, Sampaio, A.H, Cavada, B.S. | | Deposit date: | 2009-03-20 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for both pro- and anti-inflammatory response induced by mannose-specific legume lectin from Cymbosema roseum
Biochimie, 2011
|
|
3CJE
 
 | |
3CEA
 
 | |
3CU2
 
 | |
3D00
 
 | |
4Z8C
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome bound to translation inhibitor oncocin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Roy, R.N, Lomakin, I.B, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3CJM
 
 | |
3CLO
 
 | |
