5VVJ
 
 | | Cas1-Cas2 bound to half-site intermediate | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (112-MER), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.W, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
7WCA
 
 | | CATPO mutant - E484A | | Descriptor: | CALCIUM ION, Catalase, PENTAETHYLENE GLYCOL, ... | | Authors: | Yuzugullu Karakus, Y, Balci Unver, S, Zengin Karatas, M, Goc, G, Pearson, A.R, Yorke, B. | | Deposit date: | 2021-12-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Investigation of how gate residues in the main channel affect the catalytic activity of Scytalidium thermophilum catalase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7DH1
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N2-Methylguanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-12 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DPK
 
 | |
7DQN
 
 | |
7DLC
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N1-methylguanosine | | Descriptor: | 2-azanyl-9-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1-methyl-purin-6-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7DM6
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase complexed with crotonoside | | Descriptor: | 6-azanyl-9-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1H-purin-2-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DM5
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with guanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DOX
 
 | |
7DOY
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in complex with 6-O-methylguanosine | | Descriptor: | (2R,3R,4S,5R)-2-(2-azanyl-6-methoxy-purin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7DGC
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with 2'-O-Methylguanosine | | Descriptor: | 9-[(2R,3R,4R)-5-(hydroxymethyl)-3-methoxy-4-oxidanyl-oxolan-2-yl]-3H-purine-2,6-dione, Guanosine deaminase, SODIUM ION, ... | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DOW
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with 7-deazaguansoine | | Descriptor: | 2-azanyl-7-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-3H-pyrrolo[2,3-d]pyrimidin-4-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
6OGN
 
 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SGC8158 chemical probe | | Descriptor: | 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, Protein arginine N-methyltransferase 7, UNKNOWN ATOM OR ION, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response.
Nat Commun, 11, 2020
|
|
6Q6Y
 
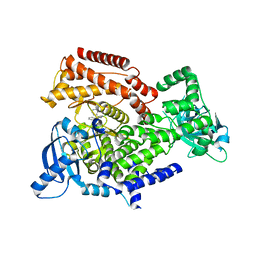 | | PI3K delta in complex with N(2chloro5phenylpyridin3yl)benzenesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-(2-chloranyl-5-phenyl-pyridin-3-yl)benzenesulfonamide | | Authors: | Convery, M.A, Rowland, P, Down, K, Barton, N. | | Deposit date: | 2018-12-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Potent, Efficient, and Selective Inhibitors of Phosphoinositide 3-Kinase delta through a Deconstruction and Regrowth Approach.
J.Med.Chem., 61, 2018
|
|
7VSQ
 
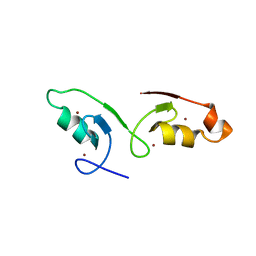 | |
7VC8
 
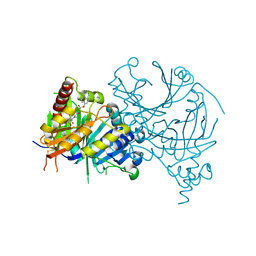 | | Complex structure of AtHPPD with inhibitor PYQ3 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, pyren-1-yl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate | | Authors: | Yang, G.F, Lin, H.Y. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7UJJ
 
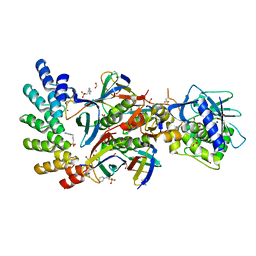 | | Stx2a and DARPin complex | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, DARPin, ... | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2022-03-30 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | A Multi-Specific DARPin Potently Neutralizes Shiga Toxin 2 via Simultaneous Modulation of Both Toxin Subunits.
Bioengineering (Basel), 9, 2022
|
|
7VSP
 
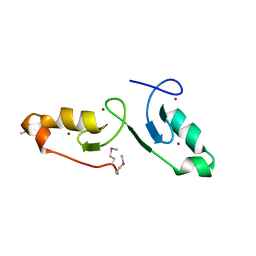 | | Crystal strcuture of the tandem B-Box domains of COL2 | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS-LIKE 2 | | Authors: | Lv, X, Liu, R, Du, J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation.
J Integr Plant Biol, 64, 2022
|
|
7WJ8
 
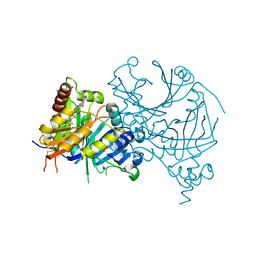 | | Complex structure of AtHPPD-PyQ1 | | Descriptor: | 2-pyren-1-yloxyethyl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-01-05 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7WJJ
 
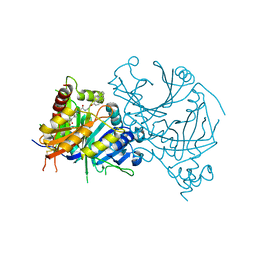 | | Complex structure of AtHPPD-PyQ2 | | Descriptor: | 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]-N-(2-pyren-1-yloxyethyl)ethanamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-01-06 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7CRA
 
 | |
7CR9
 
 | |
6OEB
 
 | | Crystal structure of HMCES SRAP domain in complex with 3' overhang DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Ravichandran, M, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OOV
 
 | | Crystal structure of HMCES SRAP domain in complex with palindromic 3' overhang DNA | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*GP*TP*TP*GP*TP*TP*TP*TP*T)-3'), Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-23 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of HMCES SRAP domain in complex with palindromic 3' overhang DNA
To Be Published
|
|
6OEA
 
 | | Crystal structure of HMCES SRAP domain in complex with longer 3' overhang DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*TP*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Ravichandran, M, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
